Search Count: 18
 |
Structure Of The Non-Canonical Ctlh E3 Substrate Receptor Wdr26 Bound To Ypel5
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-05-15 Classification: LIGASE Ligands: ZN |
 |
Structure Of The Non-Canonical Ctlh E3 Substrate Receptor Wdr26 Bound To Nmnat1 Substrate
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-05-15 Classification: LIGASE Ligands: NMN, ZN |
 |
Catalytic Module Of Human Ctlh E3 Ligase Bound To Multiphosphorylated Ube2H~Ubiquitin
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-01-03 Classification: LIGASE Ligands: ZN |
 |
Catalytic Module Of Yeast Gid E3 Ligase Bound To Multiphosphorylated Ubc8~Ubiquitin
Organism: Saccharomyces cerevisiae, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-01-03 Classification: LIGASE Ligands: ZN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2021-09-15 Classification: LIGASE Ligands: ZN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2021-09-15 Classification: LIGASE Ligands: ZN |
 |
Substrate Receptor Scaffolding Module Of Yeast Chelator-Gid Sr4 E3 Ubiquitin Ligase Bound To Fbp1 Substrate
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c), Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2021-05-05 Classification: LIGASE |
 |
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: ELECTRON MICROSCOPY Release Date: 2021-05-05 Classification: LIGASE Ligands: ZN |
 |
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2021-05-05 Classification: HYDROLASE Ligands: PO4, MG |
 |
Supramolecular Assembly Module Of Yeast Chelator-Gid Sr4 E3 Ubiquitin Ligase
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: ELECTRON MICROSCOPY Release Date: 2021-05-05 Classification: LIGASE |
 |
 |
Structure Of Active Gid E3 Ubiquitin Ligase Complex Minus Gid2 And Delta Gid9 Ring Domain
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c), Saccharomyces cerevisiae s288c, Saccharomyces cerevisiae s288c
Method: ELECTRON MICROSCOPY Release Date: 2019-11-20 Classification: LIGASE |
 |
Organism: Enterobacteria phage t4, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2017-05-24 Classification: LIGASE Ligands: 8Z7 |
 |
Organism: Enterobacteria phage t4, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.37 Å Release Date: 2017-05-24 Classification: HYDROLASE Ligands: 8ZA |
 |
Organism: Enterobacteria phage t4, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2017-05-24 Classification: Ligase / protein binding Ligands: 8ZD |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2017-05-24 Classification: Ligase / protein binding |
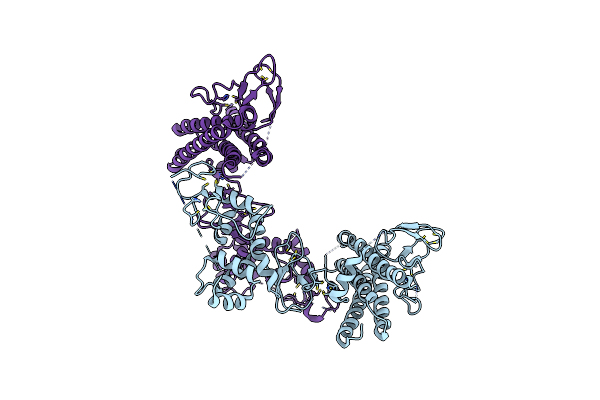 |
Structure Of Hhari, A Ring-Ibr-Ring Ubiquitin Ligase: Autoinhibition Of An Ariadne-Family E3 And Insights Into Ligation Mechanism
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.30 Å Release Date: 2013-05-29 Classification: LIGASE Ligands: ZN |
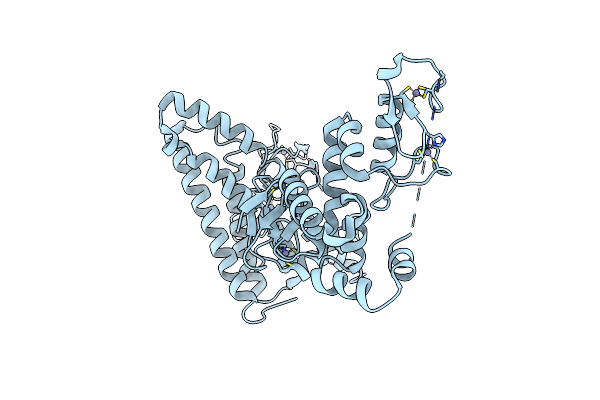 |
Structure Of Hhari, A Ring-Ibr-Ring Ubiquitin Ligase: Autoinhibition Of An Ariadne-Family E3 And Insights Into Ligation Mechanism
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.60 Å Release Date: 2013-05-29 Classification: LIGASE Ligands: ZN |

