Search Count: 147
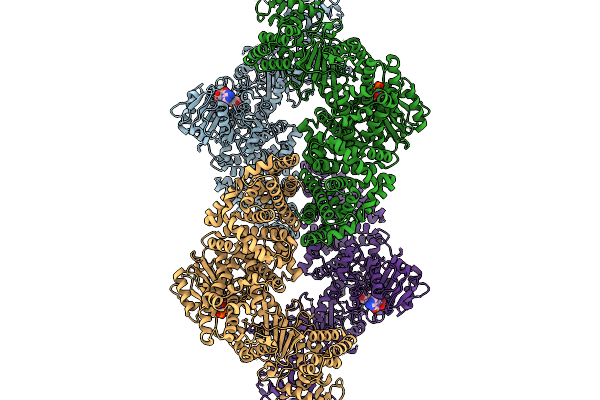 |
Cryoem Map Of The Large Glutamate Dehydrogenase Composed Of 180 Kda Subunits From Mycobacterium Smegmatis Obtained In The Presence Of Nad+ And L-Glutamate. Open Tetramer.
Organism: Mycolicibacterium smegmatis
Method: ELECTRON MICROSCOPY Release Date: 2026-01-14 Classification: OXIDOREDUCTASE Ligands: NAD |
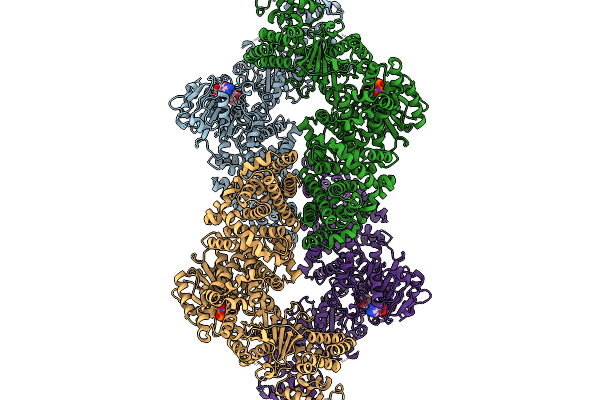 |
Cryoem Map Of The Large Glutamate Dehydrogenase Composed Of 180 Kda Subunits From Mycobacterium Smegmatis Obtained In The Presence Of Nad+ And L-Glutamate. Closed1 Tetramer.
Organism: Mycolicibacterium smegmatis
Method: ELECTRON MICROSCOPY Release Date: 2026-01-14 Classification: OXIDOREDUCTASE Ligands: NAD |
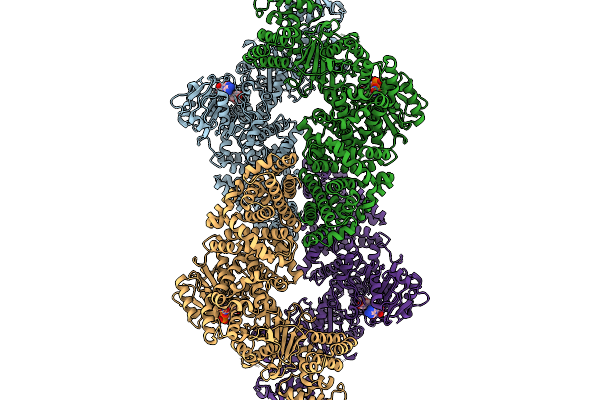 |
Cryoem Map Of The Large Glutamate Dehydrogenase Composed Of 180 Kda Subunits From Mycobacterium Smegmatis Obtained In The Presence Of Nad+ And L-Glutamate. Closed2 Tetramer
Organism: Mycolicibacterium smegmatis
Method: ELECTRON MICROSCOPY Release Date: 2026-01-14 Classification: OXIDOREDUCTASE Ligands: NAD |
 |
Cryoem Map Of The Large Glutamate Dehydrogenase Composed Of 180 Kda Subunits From Mycobacterium Smegmatis Obtained In The Presence Of Nad+ And L-Glutamate. Empty Monomer.
Organism: Mycolicibacterium smegmatis
Method: ELECTRON MICROSCOPY Release Date: 2026-01-14 Classification: OXIDOREDUCTASE |
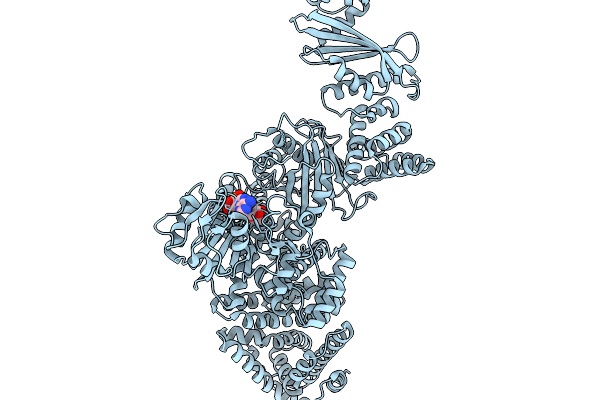 |
Cryoem Map Of The Large Glutamate Dehydrogenase Composed Of 180 Kda Subunits From Mycobacterium Smegmatis Obtained In The Presence Of Nad+ And L-Glutamate. Cofactor-Monomer.
Organism: Mycolicibacterium smegmatis
Method: ELECTRON MICROSCOPY Release Date: 2026-01-14 Classification: OXIDOREDUCTASE Ligands: NAD |
 |
Cryoem Map Of The Large Glutamate Dehydrogenase Composed Of 180 Kda Subunits From Mycobacterium Smegmatis Obtained In The Presence Of Nad+ And L-Glutamate. Cofactor/Ligand-Monomer
Organism: Mycolicibacterium smegmatis
Method: ELECTRON MICROSCOPY Release Date: 2026-01-14 Classification: OXIDOREDUCTASE Ligands: GLU, NAD |
 |
Cryoem Map Of The Large Glutamate Dehydrogenase Composed Of 180 Kda Subunits From Mycobacterium Smegmatis Obtained In The Presence Of Nad+ And L-Glutamate. Total-Monomer
Organism: Mycolicibacterium smegmatis
Method: ELECTRON MICROSCOPY Release Date: 2026-01-14 Classification: OXIDOREDUCTASE Ligands: NAD |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: CHAPERONE Ligands: ATP, MG, K |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: CHAPERONE Ligands: ATP, MG, K |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: CHAPERONE Ligands: ATP, MG, K |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: CHAPERONE Ligands: ATP, MG, K |
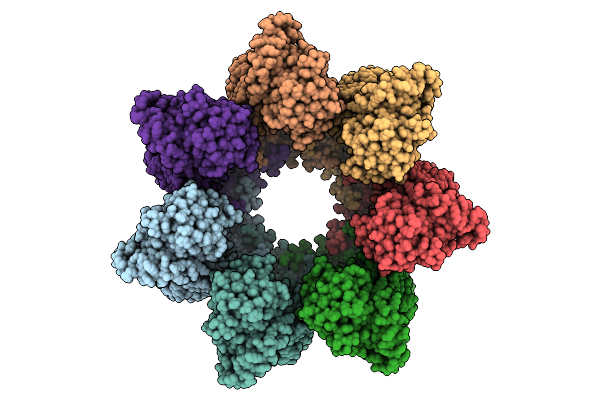 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: CHAPERONE Ligands: ADP, MG |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: CHAPERONE Ligands: ATP, MG, K |
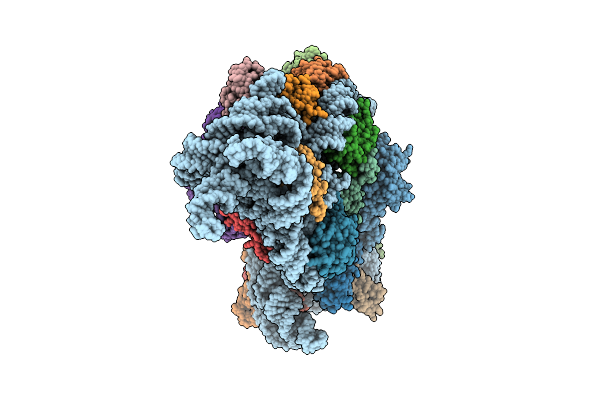 |
Bacterial 30S Ribosomal Subunit Assembly Complex State B (Consensus Refinement)
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2025-11-12 Classification: RIBOSOME Ligands: MG, ZN |
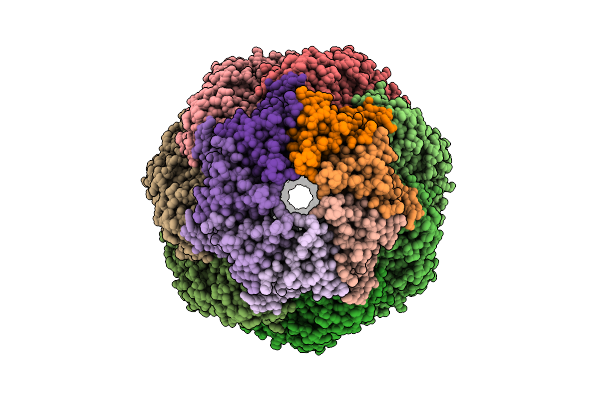 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: CHAPERONE Ligands: ATP, MG, K |
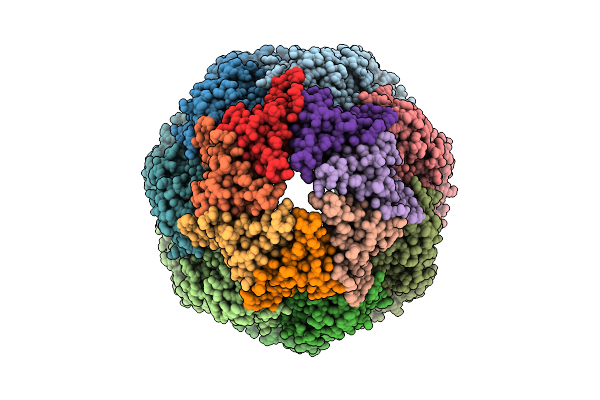 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: CHAPERONE Ligands: ATP, MG, K |
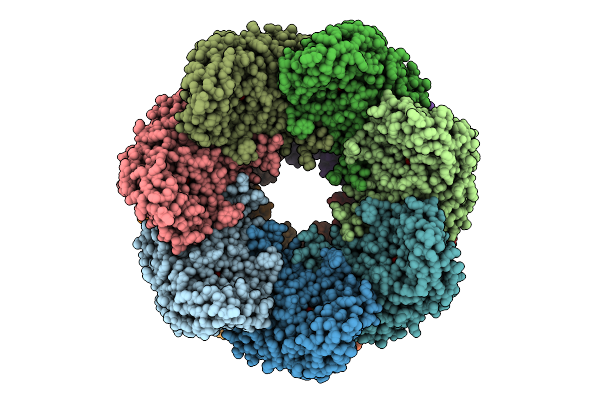 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: CHAPERONE Ligands: ATP, MG, K |
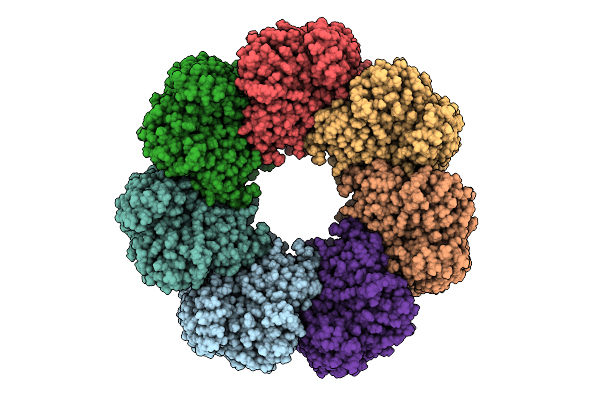 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: CHAPERONE |
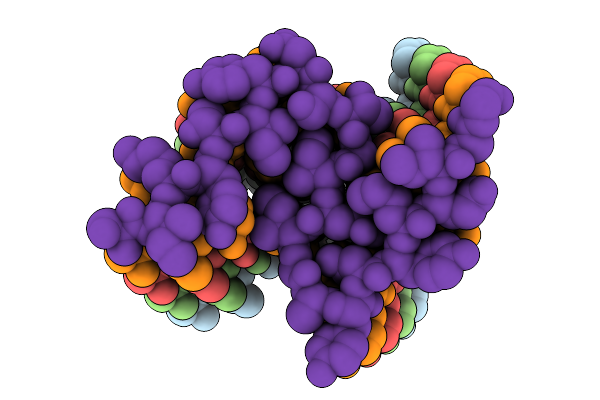 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-09-17 Classification: PROTEIN FIBRIL |
 |
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2025-08-06 Classification: RIBOSOME Ligands: A1IC4, MG, K |

