Search Count: 13
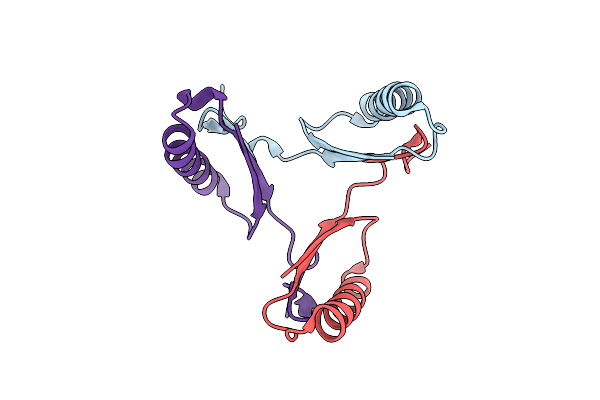 |
Crystal Structure Of Dmpi From Archaeoglobus Fulgidus Determined To 2.37 Angstroms Resolution
Organism: Archaeoglobus fulgidus
Method: X-RAY DIFFRACTION Resolution:2.37 Å Release Date: 2010-09-01 Classification: ISOMERASE |
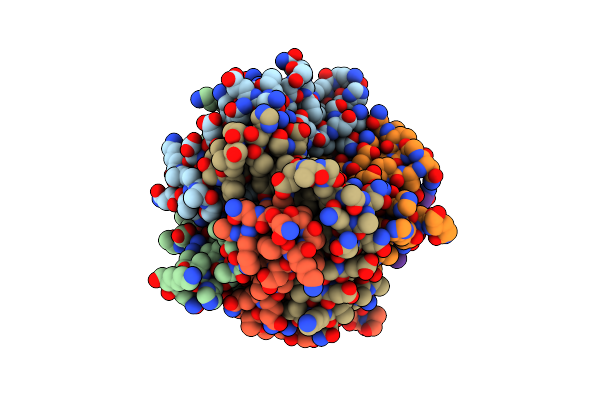 |
Crystal Structure Of Dmpi From Helicobacter Pylori Determined To 1.9 Angstroms Resolution
Organism: Helicobacter pylori
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2010-09-01 Classification: ISOMERASE |
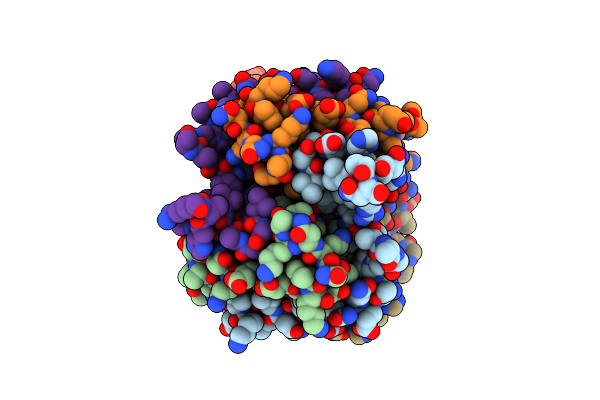 |
Crystal Structure Of The 4-Oxalocrotonate Tautomerase Homologue Dmpi From Helicobacter Pylori.
Organism: Helicobacter pylori
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2008-02-12 Classification: ISOMERASE |
 |
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2008-02-05 Classification: ISOMERASE |
 |
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2008-02-05 Classification: ISOMERASE Ligands: FHC |
 |
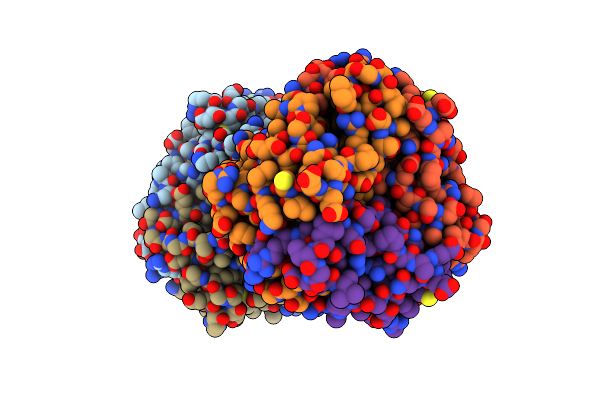
Evolution Of Enzymatic Activity In The Tautomerase Superfamily: Mechanistic And Structural Consequences Of The L8R Mutation In 4-Oxalocrotonate Tautomerase
Organism: Pseudomonas putida
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2006-09-26 Classification: TRANSFERASE Ligands: CL |
 |
Crystal Structures Of The Wild-Type, Mutant-P1A And Inactivated Malonate Semialdehyde Decarboxylase: A Structural Basis For The Decarboxylase And Hydratase Activities
Organism: Pseudomonas pavonaceae
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2005-11-22 Classification: LYASE |
 |
Crystal Structures Of The Wild-Type, Mutant-P1A And Inactivated Malonate Semialdehyde Decarboxylase: A Structural Basis For The Decarboxylase And Hydratase Activities
Organism: Pseudomonas pavonaceae
Method: X-RAY DIFFRACTION Resolution:2.74 Å Release Date: 2005-11-22 Classification: LYASE |
 |
Crystal Structures Of The Wild-Type, Mutant-P1A And Inactivated Malonate Semialdehyde Decarboxylase: A Structural Basis For The Decarboxylase And Hydratase Activities
Organism: Pseudomonas pavonaceae
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2005-11-22 Classification: LYASE Ligands: MLI |
 |
The Crystal Structure Of Ydce, A 4-Oxalocrotonate Tautomerase Homologue From Escherichia Coli, Confirms The Structural Basis For Oligomer Diversity
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2002-10-10 Classification: ISOMERASE |
 |
The Crystal Structure Of Ydce, A 4-Oxalocrotonate Tautomerase Homologue From Escherichia Coli, Confirms The Structural Basis For Oligomer Diversity
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2002-10-10 Classification: ISOMERASE Ligands: BEZ, EPE |
 |
The Crystal Structure Of Ydce, A 4-Oxalocrotonate Tautomerase Homologue From Escherichia Coli, Confirms The Structural Basis For Oligomer Diversity
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2002-10-10 Classification: ISOMERASE Ligands: FHC |
 |
Crystal Structure Of Human Ornithine Decarboxylase At 2.1 Angstroms Resolution
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2000-10-25 Classification: LYASE |

