Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
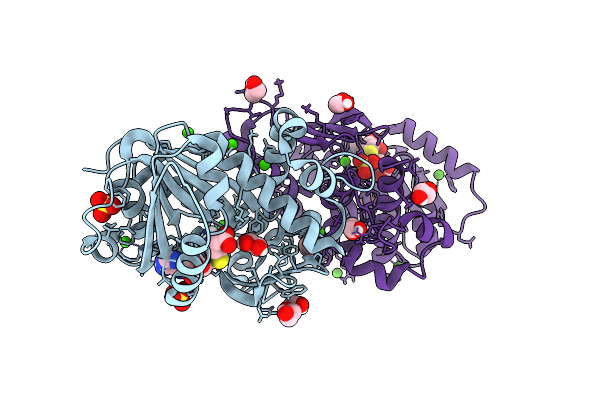 |
Structure Of Kluyveromyces Lactis Mrna Cap (Guanine-N7) Methyltransferase, Abd1, In Complex With Sah
Organism: Kluyveromyces lactis nrrl y-1140
Method: X-RAY DIFFRACTION Release Date: 2025-07-09 Classification: TRANSFERASE Ligands: SAH, EDO, PEG, SO4, CA |
 |
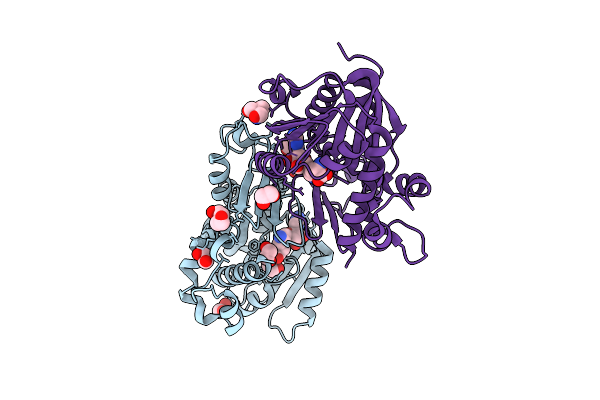
Structure Of Kluyveromyces Lactis Mrna Cap (Guanine-N7) Methyltransferase, Abd1, In Complex With Adenine And M7Gtp
Organism: Kluyveromyces lactis nrrl y-1140
Method: X-RAY DIFFRACTION Release Date: 2025-07-09 Classification: TRANSFERASE Ligands: ADE, MGP, P4G, SO4, EDO |
 |
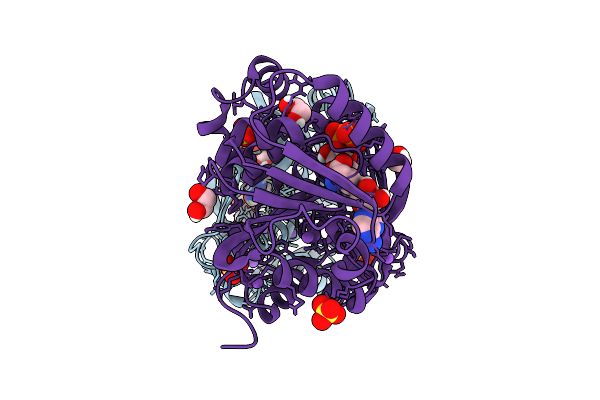
Structure Of Kluyveromyces Lactis Mrna Cap (Guanine-N7) Methyltransferase, Abd1, In Complex With Sinefungin
Organism: Kluyveromyces lactis nrrl y-1140
Method: X-RAY DIFFRACTION Release Date: 2025-07-09 Classification: TRANSFERASE Ligands: SFG, EDO, PEG, TRS |
 |
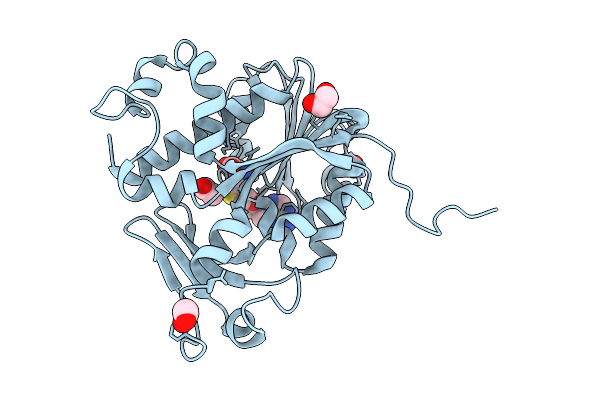
Structure Of Kluyveromyces Lactis Mrna Cap (Guanine-N7) Methyltransferase, Abd1, In Complex With Sinefungin And Gtp
Organism: Kluyveromyces lactis nrrl y-1140
Method: X-RAY DIFFRACTION Release Date: 2025-07-09 Classification: TRANSFERASE Ligands: SFG, GTP, NA, SO4, EDO |
 |
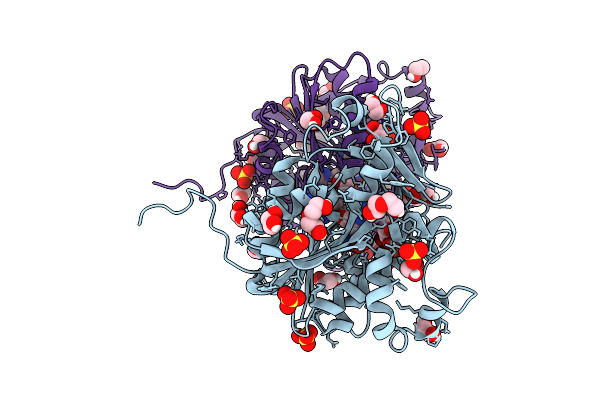
Structure Of Saccharomyces Cerevisiae Mrna Cap (Guanine-N7) Methyltransferase, Abd1, In Complex With Sah
Organism: Saccharomyces cerevisiae yjm789
Method: X-RAY DIFFRACTION Release Date: 2025-07-09 Classification: TRANSFERASE Ligands: SAH, EDO, ACT |
 |
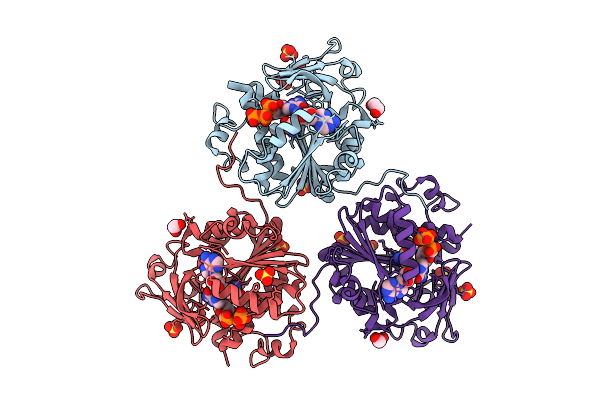
Structure Of Saccharomyces Cerevisiae Mrna Cap (Guanine-N7) Methyltransferase, Abd1, In Complex With Sinefungin And Gtp
Organism: Saccharomyces cerevisiae yjm789
Method: X-RAY DIFFRACTION Release Date: 2025-07-09 Classification: TRANSFERASE Ligands: SFG, GTP, PEG, EDO, SO4, 1PE, CL |
 |
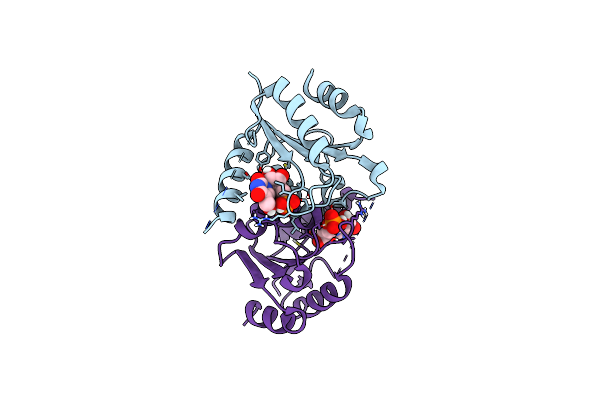
Structure Of Saccharomyces Cerevisiae Mrna Cap (Guanine-N7) Methyltransferase Variant, Abd1-K163R-K311R-F387Y-Y416F, In Complex With Sinefungin And Gtp
Organism: Saccharomyces cerevisiae yjm789
Method: X-RAY DIFFRACTION Release Date: 2025-07-09 Classification: TRANSFERASE Ligands: SFG, GTP, EDO, SO4 |
 |
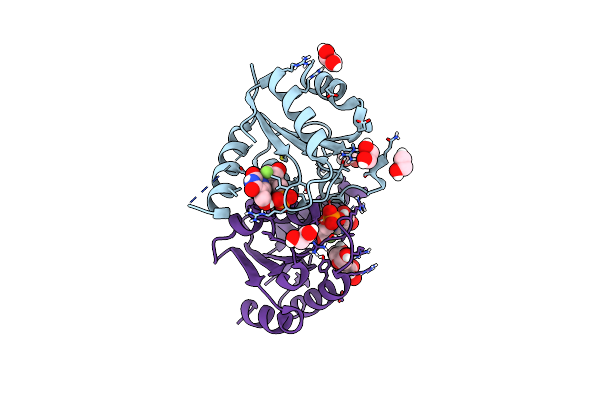
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.47 Å Release Date: 2025-02-05 Classification: HYDROLASE/INHIBITOR Ligands: A1BBB |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.13 Å Release Date: 2025-02-05 Classification: HYDROLASE/INHIBITOR Ligands: A1BBC, EDO, PEG |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.51 Å Release Date: 2025-02-05 Classification: HYDROLASE/INHIBITOR Ligands: A1BBD |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.73 Å Release Date: 2025-02-05 Classification: HYDROLASE/INHIBITOR Ligands: A1BBE |
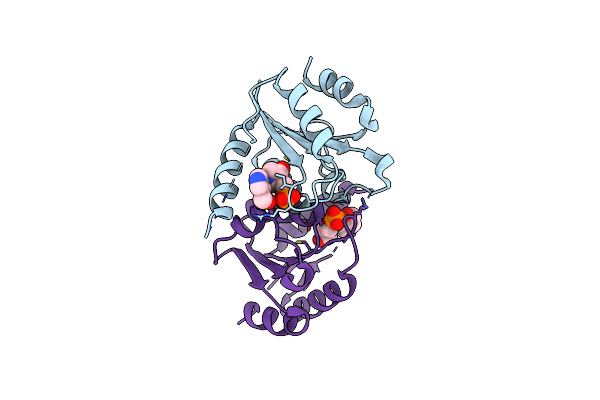 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.82 Å Release Date: 2025-02-05 Classification: HYDROLASE/INHIBITOR Ligands: A1BBF |
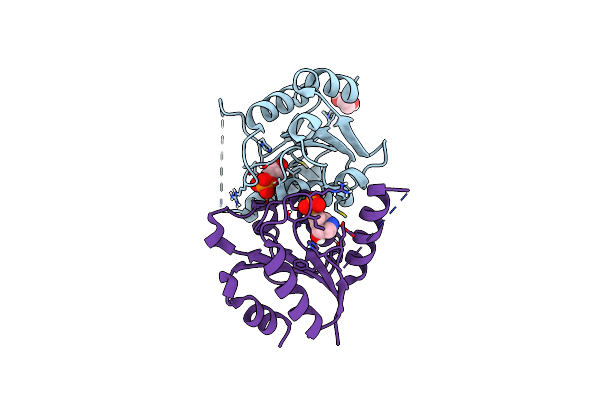 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2025-02-05 Classification: HYDROLASE/INHIBITOR Ligands: NR1, EDO |
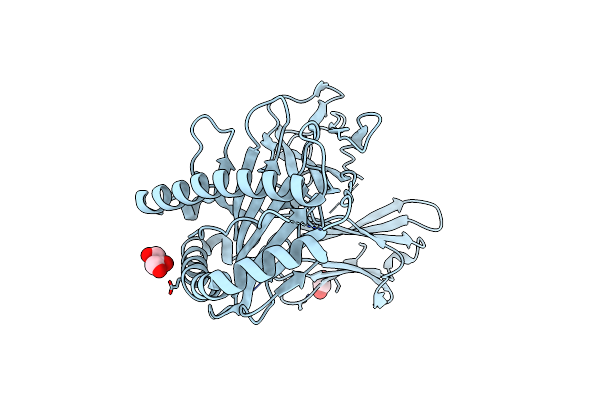 |
Crystal Structure Of A Single Chain Trimer Composed Of Hla-B*39:06 Y84C Variant, Beta-2Microglobulin, And Nrvmlpkaa Peptide From Nlrp2 (1 Molecule/Asymmetric Unit)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2024-09-04 Classification: IMMUNE SYSTEM Ligands: EDO, GOL, CL |
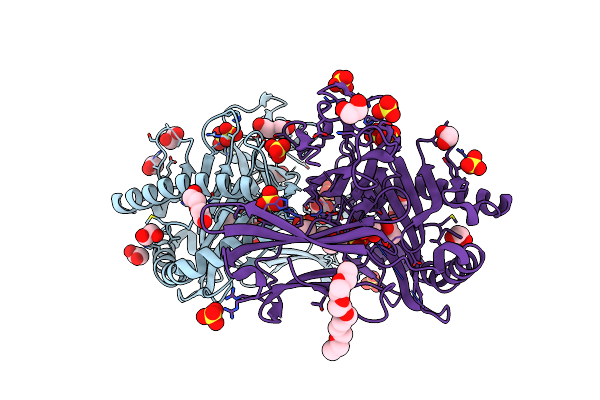 |
Crystal Structure Of A Single Chain Trimer Composed Of Hla-B*39:06 Y84C Variant, Beta-2Microglobulin, And Nrvmlpkaa Peptide From Nlrp2 (2 Molecules/Asymmetric Unit)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2024-09-04 Classification: IMMUNE SYSTEM Ligands: SO4, EDO, PEG, CL, P3G, GOL |
 |
Crystal Structure Of A Single Chain Trimer Composed Of Hla-B*39:01 Y84C Variant, Beta-2Microglobulin, And Nrvmlpkaa Peptide From Nlrp2
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2024-09-04 Classification: IMMUNE SYSTEM Ligands: P4G, EDO |
 |
Structure Of The Mena Evh1 Domain Bound To The Polyproline Segment Of Ptp1B
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2024-08-28 Classification: STRUCTURAL PROTEIN Ligands: EDO, SO4 |
 |
Organism: Aequorea victoria, Lama glama
Method: X-RAY DIFFRACTION Release Date: 2024-05-22 Classification: IMMUNE SYSTEM Ligands: NH4, GOL, CL, SO4, NA |
 |
Organism: Aequorea victoria, Lama glama
Method: X-RAY DIFFRACTION Release Date: 2024-05-22 Classification: IMMUNE SYSTEM Ligands: GOL, SO4, NA |
 |
Organism: Aequorea victoria, Lama glama
Method: X-RAY DIFFRACTION Release Date: 2024-05-22 Classification: IMMUNE SYSTEM Ligands: GOL, MLT, NA, PO4 |