Search Count: 50
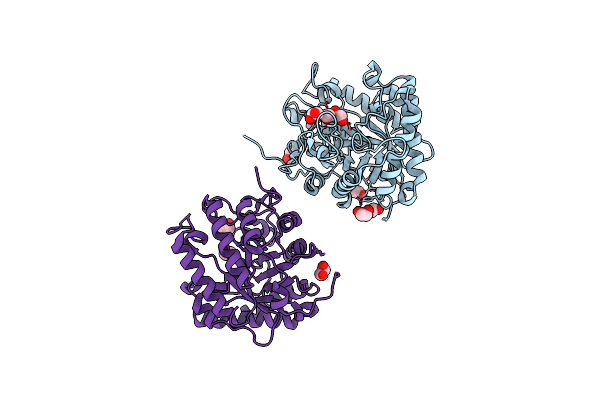 |
Crystal Structure Of Celd Cellulase From The Anaerobic Fungus Piromyces Finnis
Organism: Piromyces finnis
Method: X-RAY DIFFRACTION Resolution:2.46 Å Release Date: 2023-05-17 Classification: HYDROLASE Ligands: EDO |
 |
Crystal Structure Of The E154D Mutant Celd Cellulase From The Anaerobic Fungus Piromyces Finnis In The Complex With Cellotriose.
Organism: Piromyces finnis
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2023-05-17 Classification: HYDROLASE |
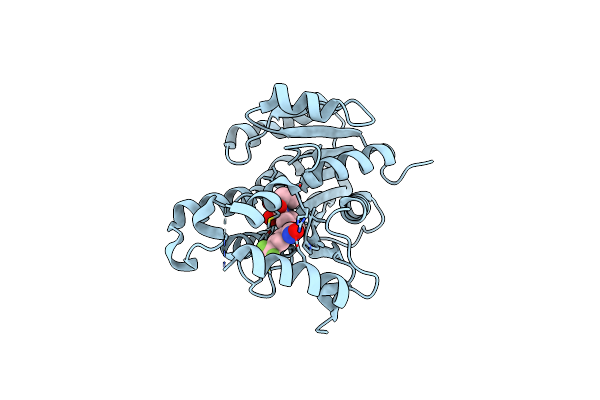 |
Crystal Structure Of The Inha From Mycobacterium Tuberculosis In Complex With An12855, Ebsi 4333.
Organism: Mycobacterium tuberculosis (strain cdc 1551 / oshkosh)
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2018-06-13 Classification: OXIDOREDUCTASE Ligands: 9JA |
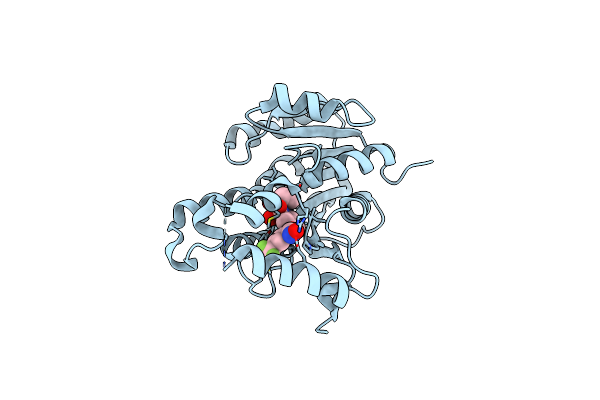 |
Crystal Structure Of The Inha From Mycobacterium Tuberculosis In Complex With An12855, Ebsi 4333.
Organism: Mycobacterium tuberculosis (strain cdc 1551 / oshkosh)
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2018-05-16 Classification: OXIDOREDUCTASE Ligands: 9JA |
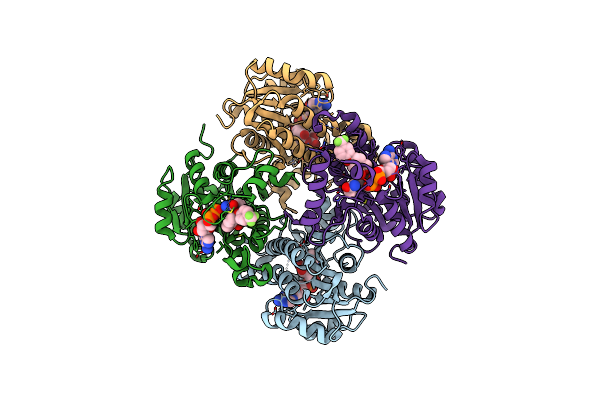 |
Crystal Structure Of The Inha From Mycobacterium Tuberculosis In Complex With An12855, Ebsi 4333.
Organism: Mycobacterium tuberculosis (strain cdc 1551 / oshkosh)
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2018-05-16 Classification: OXIDOREDUCTASE Ligands: 9JJ |
 |
Crystal Structure Of The Inha From Mycobacterium Tuberculosis In Complex With An12855, Ebsi 4333.
Organism: Mycobacterium tuberculosis (strain cdc 1551 / oshkosh)
Method: X-RAY DIFFRACTION Resolution:2.55 Å Release Date: 2018-05-16 Classification: OXIDOREDUCTASE Ligands: 9JM |
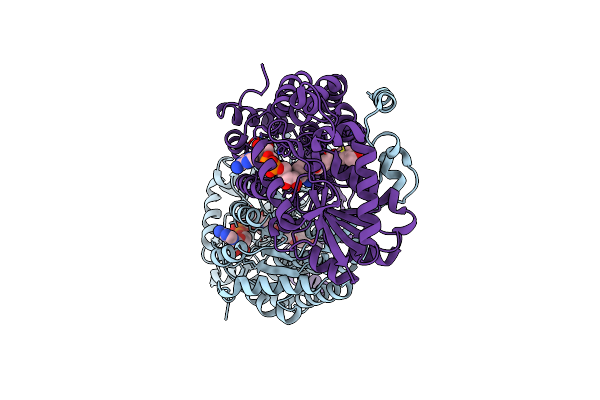 |
Organism: Trypanosoma brucei
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2017-04-19 Classification: OXIDOREDUCTASE Ligands: NAD, 92N |
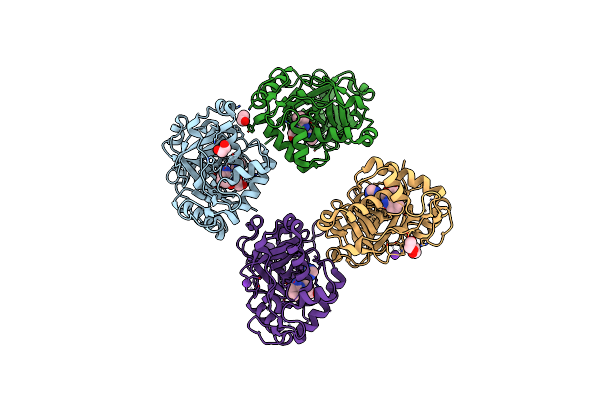 |
Crystal Structure Of Hte Cryptosporidium Muris Cytosolic Leucyl-Trna Synthetase Editing Domain Complex With A Post-Transfer Editing Analogue Of Norvaline (Nv2Aa)
Organism: Cryptosporidium muris
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2016-08-03 Classification: LYASE Ligands: VRT, EDO, K |
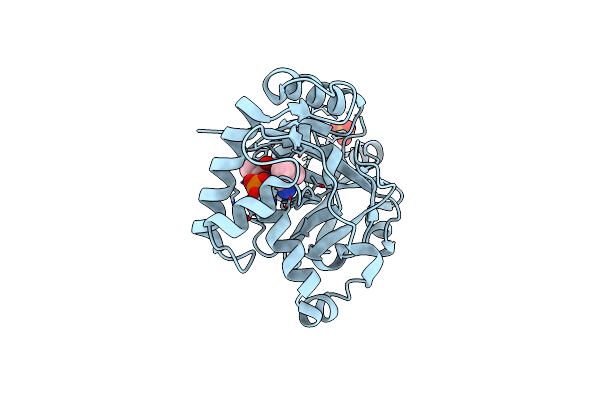 |
Crystal Structure Of The Cryptosporidium Muris Cytosolic Leucyl-Trna Synthetase Editing Domain Complex With A Post-Transfer Editing Analogue Of Isoeucine (Ile2Aa)
Organism: Cryptosporidium muris
Method: X-RAY DIFFRACTION Resolution:1.77 Å Release Date: 2016-08-03 Classification: LYASE Ligands: PO4, SO8 |
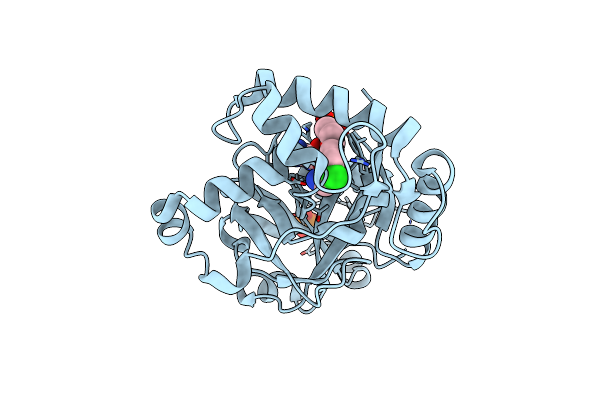 |
Crystal Structure Of The Cryptosporidium Muris Cytosolic Leucyl-Trna Synthetase Editing Domain Complex With The Adduct Amp-An6426
Organism: Cryptosporidium muris
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2016-08-03 Classification: LIGASE Ligands: A2H, PO4 |
 |
Crystal Structure Of The Cryptosporidium Muris Cytosolic Leucyl-Trna Synthetase Editing Domain (Apo Structure)
Organism: Cryptosporidium muris
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2016-08-03 Classification: LIGASE |
 |
Crystal Structure Of The P.Falciparum Cytosolic Leucyl-Trna Synthetase Editing Domain (Space Group P1)
Organism: Plasmodium falciparum
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2016-06-22 Classification: LIGASE |
 |
Crystal Structure Of The P.Falciparum Cytosolic Leucyl-Trna Synthetase Editing Domain (Space Group P21)
Organism: Plasmodium falciparum
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2016-06-22 Classification: LIGASE |
 |
Crystal Structure Of The P.Falciparum Cytosolic Leucyl-Trna Synthetase Editing Domain (Space Group P1) Containing Deletions Of Insertions 1 And 3
Organism: Plasmodium falciparum
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2016-06-22 Classification: LIGASE Ligands: EDO |
 |
Crystal Structure Of The P.Knowlesi Cytosolic Leucyl-Trna Synthetase Editing Domain
Organism: Plasmodium knowlesi
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2016-06-22 Classification: LIGASE |
 |
Crystal Structure Of The Leurs Editing Domain Of Mycobacterium Tuberculosis In Complex With The Adduct 3-(Aminomethyl)-4-Bromo-7-Ethoxybenzo[C][1,2]Oxaborol-1(3H)-Ol-Amp
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:1.47 Å Release Date: 2016-03-09 Classification: LIGASE Ligands: MET, 38Y |
 |
Crystal Structure Of The Leurs Editing Domain Of Mycobacterium Tuberculosis In Complex With The Adduct (S)-3-(Aminomethyl)-7-Ethoxybenzo[C][1,2]Oxaborol-1(3H)-Ol-Amp
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2016-03-02 Classification: LIGASE Ligands: MET, LEU, A52, EDO |
 |
Crystal Structure Of The Leurs Editing Domain Of Mycobacterium Tuberculosis In Complex With The Adduct (S)-3-(Aminomethyl)-4-Chloro-7-Ethoxybenzo[C][1,2]Oxaborol-1(3H)-Ol-Amp
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2016-03-02 Classification: LIGASE Ligands: MET, A2H, GOL |
 |
Ternary Complex Of E.Coli Leucyl-Trna Synthetase, Trna(Leu)574 And The Benzoxaborole An3017 In The Editing Conformation
Organism: Escherichia coli, Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2013-04-17 Classification: LIGASE/RNA Ligands: MG |
 |
Ternary Complex Of E .Coli Leucyl-Trna Synthetase, Trna(Leu) And The Benzoxaborole An3016 In The Editing Conformation
Organism: Escherichia coli, Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2013-04-17 Classification: LIGASE/RNA Ligands: MG |

