Search Count: 10
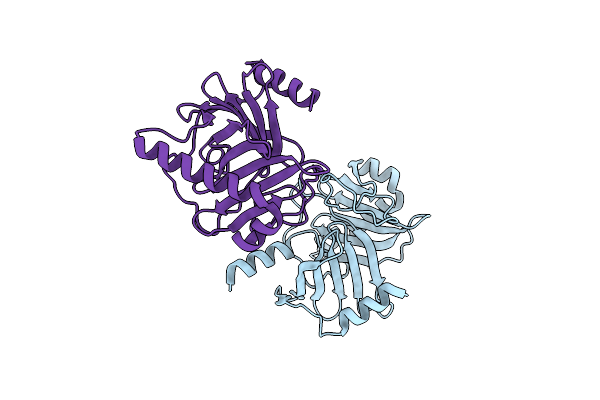 |
Crystal Structure Of The Kinetoplastid Kinetochore Protein Trypanosoma Congolense Kkt2 Divergent Polo-Box Domain
Organism: Trypanosoma congolense il3000
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2022-06-15 Classification: CELL CYCLE |
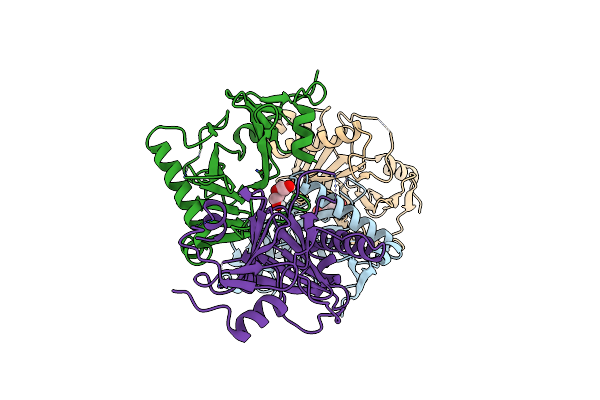 |
Crystal Structure Of The Kinetoplastid Kinetochore Protein Trypanosoma Brucei Kkt3 Divergent Polo-Box Domain
Organism: Trypanosoma brucei brucei
Method: X-RAY DIFFRACTION Resolution:2.92 Å Release Date: 2022-06-15 Classification: CELL CYCLE Ligands: PEG |
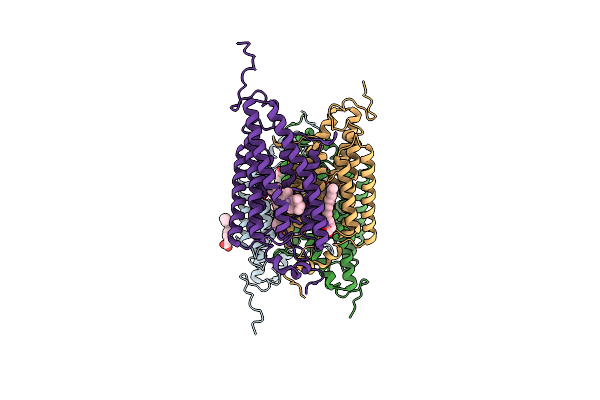 |
Organism: Guillardia theta ccmp2712
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2018-09-05 Classification: MEMBRANE PROTEIN Ligands: RET, OLA |
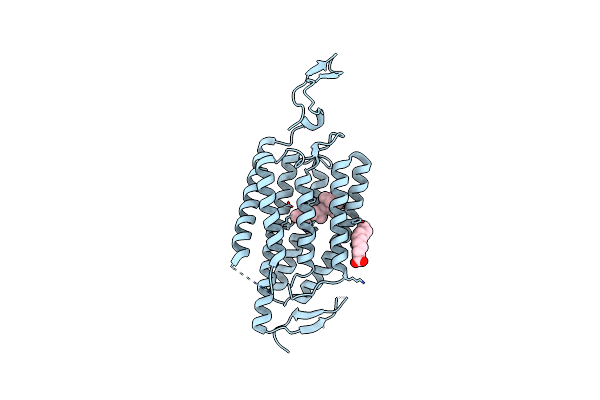 |
Organism: Chlamydomonas reinhardtii
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2018-09-05 Classification: MEMBRANE PROTEIN Ligands: RET, OLA, CL |
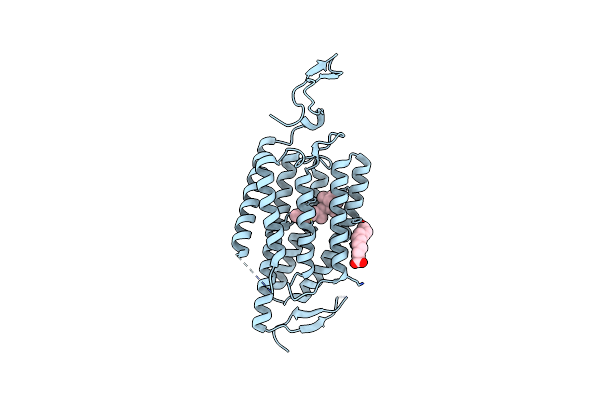 |
Organism: Chlamydomonas reinhardtii
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2018-09-05 Classification: MEMBRANE PROTEIN Ligands: RET, OLA |
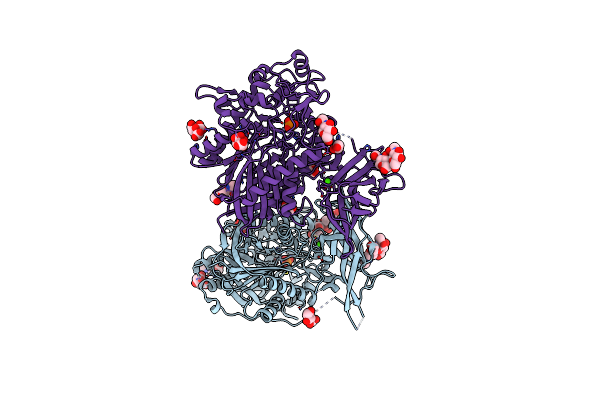 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.58 Å Release Date: 2015-07-08 Classification: HYDROLASE Ligands: ZN, CA, NAG, PO4, CIT, CL, EDO |
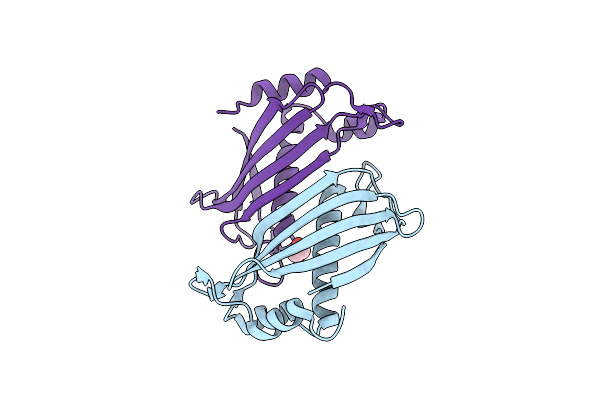 |
Crystal Structure Of Slr0204, A 1,4-Dihydroxy-2-Naphthoyl-Coa Thioesterase From Synechocystis
Organism: Synechocystis sp.
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2013-04-17 Classification: HYDROLASE Ligands: EDO |
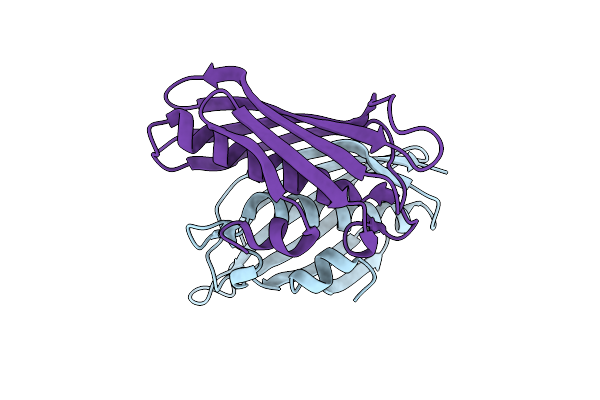 |
Crystal Structure Of Atdhnat1, A 1,4-Dihydroxy-2-Naphthoyl-Coa Thioesterase From Arabidopsis Thaliana
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2013-04-17 Classification: HYDROLASE |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:3.01 Å Release Date: 2011-06-01 Classification: MEMBRANE PROTEIN |
 |
Crystal Structure Of The Fimd Usher Bound To Its Cognate Fimc:Fimh Substrate
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2011-06-01 Classification: CELL ADHESION/TRANSPORT/CHAPERONE Ligands: SO4 |

