Search Count: 45
 |
From Dna-Encoded Library Screening To Am-9747 - An Mta-Cooperative Prmt5 Inhibitor With Potent Oral In Vivo Efficacy
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2025-03-26 Classification: TRANSFERASE/INHIBITOR Ligands: A1BQW, MTA, GOL, PEG, DMS |
 |
Crystal Structure Of Human Kras G12C Covalently Bound To Del Triazine Compound 5
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2025-02-26 Classification: SIGNALING PROTEIN,ONCOPROTEIN/INHIBITOR Ligands: CA, GDP, A1BH6 |
 |
Crystal Structure Of Human Kras G12C Covalently Bound To Nopinone-Derived Naphthol Compound 21
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.18 Å Release Date: 2025-02-26 Classification: SIGNALING PROTEIN,ONCOPROTEIN/INHIBITOR Ligands: MG, GDP, A1BH5, EDO |
 |
Amg 193, A Clinical Stage Mta-Cooperative Prmt5 Inhibitor, Drives Anti-Tumor Activity Preclinically And In Patients With Mtap-Deleted Cancers
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.85 Å Release Date: 2024-10-02 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: GOL, DMS, A1ATH, MTA |
 |
Crystal Structure Of A Mycobacteriophage Cluster A2 Immunity Repressor:Dna Complex
Organism: Mycobacterium phage tipsythetrex
Method: X-RAY DIFFRACTION Resolution:3.13 Å Release Date: 2022-07-20 Classification: GENE REGULATION/DNA |
 |
Crystal Structure Of A Mycobacteriophage Cluster A2 Immunity Repressor:Dna Complex
Organism: Mycobacterium phage tipsythetrex
Method: X-RAY DIFFRACTION Resolution:2.79 Å Release Date: 2022-07-20 Classification: GENE REGULATION/DNA |
 |
Organism: Sphingomonas sp. kt-1
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2021-12-08 Classification: HYDROLASE Ligands: ZN |
 |
Organism: Sphingomonas sp. kt-1
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2021-12-08 Classification: HYDROLASE Ligands: GD3 |
 |
A Structural Characterization Of Poly(Aspartic Acid) Hydrolase-1 From Sphingomonas Sp. Kt-1.
Organism: Sphingomonas sp. kt-1
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2020-12-09 Classification: HYDROLASE |
 |
Crystal Structure Of Bace1 In Complex With (Z)-Fluoro-Olefin Containing Compound 15
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.86 Å Release Date: 2020-06-03 Classification: HYDROLASE/HYDROLASE Inhibitor Ligands: U64, IOD |
 |
Structure Of T7 Dna Polymerase Bound To A Primer/Template Dna And A Peptide That Mimics The C-Terminal Tail Of The Primase-Helicase
Organism: Enterobacteria phage t7, Escherichia coli
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2020-03-04 Classification: TRANSFERASE/DNA BINDING PROTEIN/DNA Ligands: TTP, MG |
 |
Crystal Structure Of Human Kras G12C Covalently Bound To A Phthalazine Inhibitor
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2019-12-25 Classification: SIGNALING PROTEIN/Inhibitor Ligands: MG, GDP, OJ1 |
 |
Crystal Structure Of Human Kras G12C Covalently Bound To A Quinazolinone Inhibitor
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2019-12-25 Classification: SIGNALING PROTEIN/Inhibitor Ligands: GDP, CA, OHY |
 |
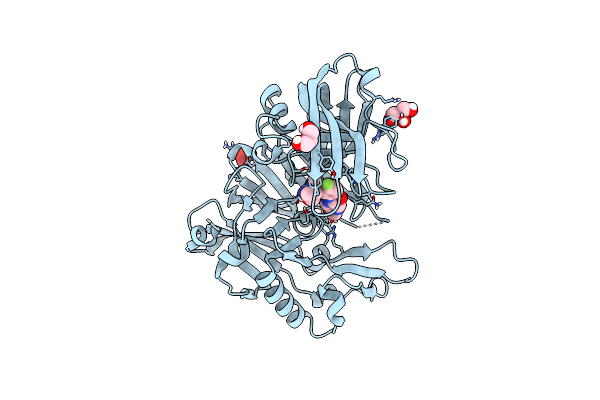
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2019-10-23 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: IOD, GOL, P6J |
 |
Structure Of Bace-1 (Beta-Secretase) In Complex With : N-(3-((1R,5S,6R)-3-Amino-5-Methyl-2-Oxa-4-Azabicyclo[4.1.0]Hept-3-En-5-Yl)-4-Fluorophenyl)-5-Methoxypyrazine-2-Carboxamide
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2018-02-21 Classification: Hydrolase/Hydrolase Inhibitor Ligands: IOD, GOL, SO4, EJ7 |
 |
Organism: Enterobacteria phage t7, Escherichia coli o157:h7
Method: X-RAY DIFFRACTION Resolution:4.80 Å Release Date: 2016-12-07 Classification: TRANSFERASE |
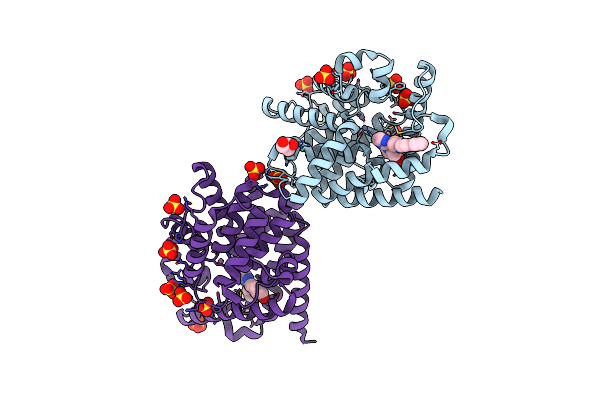 |
Crystal Structure Of 2-(3-Alkoxy-1-Azetidinyl) Quinolines As Pde10A Inhibitors
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.77 Å Release Date: 2014-12-17 Classification: Hydrolase/Hydrolase Inhibitor Ligands: SO4, GOL, 35E, ZN |
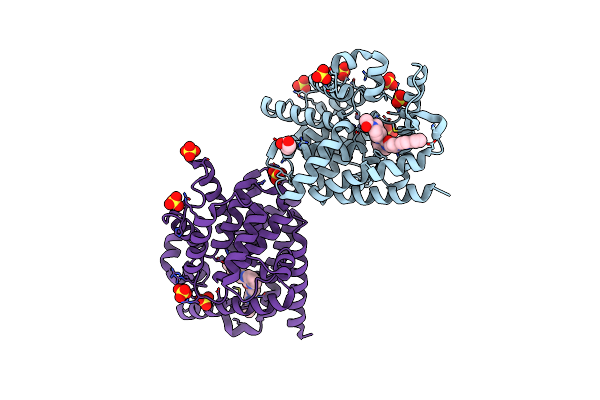 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2014-12-17 Classification: Hydrolase/Hydrolase Inhibitor Ligands: ZN, SO4, GOL, 35D |
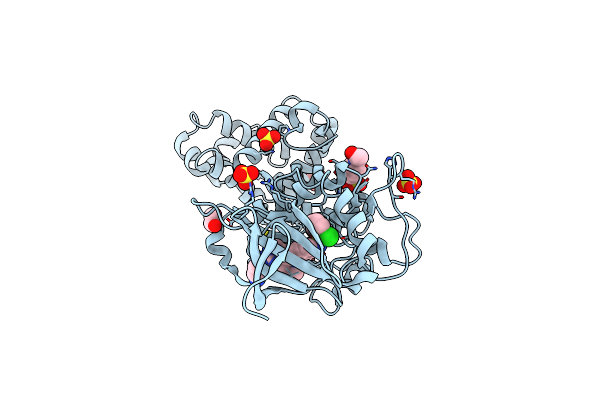 |
Crystal Structure Of Human Ire1 Cytoplasmic Domains In Complex With A Sulfonamide Inhibitor.
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2014-10-08 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: 3E4, SO4, EDO, EPE |
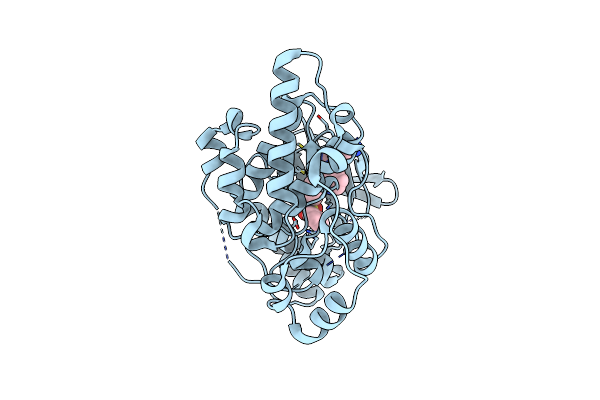 |
Crystal Structure Of Human Jnk3 In Complex With A Benzenesulfonamide Inhibitor.
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.23 Å Release Date: 2014-10-08 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: 3EL |

