Search Count: 168
 |
Solution Structure Of Risdiplam Bound To The Rna Duplex Formed Upon 5'-Splice Site Recognition
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 2024-03-06 Classification: RNA Ligands: Y59 |
 |
Solution Structure Of Branaplam Bound To The Rna Duplex Formed Upon 5'-Splice Site Recognition
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 2024-03-06 Classification: SPLICING Ligands: Y53 |
 |
Solution Structure Of Smn-Cx Bound To The Rna Helix Formed Upon Smn2 Exon7 5'-Splice Site Recognition
Organism: Homo sapiens
Method: SOLID-STATE NMR Release Date: 2024-03-06 Classification: SPLICING Ligands: YB3 |
 |
Solution Structure Of The Rna Helix Formed By The 5'-End Of U1 Snrna And An A-1 Bulged 5'-Splice Site In Complex With Smn-Cy
Organism: Synthetic construct
Method: SOLUTION NMR Release Date: 2024-02-21 Classification: SPLICING Ligands: ULR |
 |
Organism: Salmonella enterica
Method: SOLUTION NMR Release Date: 2024-01-17 Classification: TRANSPORT PROTEIN |
 |
Nmr Solution Structure Of The N-Terminal Rrm And Flanking Linker Regions Of Polypyrimidine Tract Binding Protein 1 Using The Cyana Consensus Method.
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 2023-11-08 Classification: RNA BINDING PROTEIN |
 |
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 2023-03-29 Classification: RNA BINDING PROTEIN |
 |
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 2023-02-08 Classification: RNA BINDING PROTEIN |
 |
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c), Synthetic construct
Method: SOLUTION NMR Release Date: 2022-12-21 Classification: RNA BINDING PROTEIN |
 |
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c), Synthetic construct
Method: SOLUTION NMR Release Date: 2022-10-26 Classification: RNA BINDING PROTEIN |
 |
Organism: Synthetic construct, Streptococcus pyogenes
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2022-10-26 Classification: HYDROLASE Ligands: MG, K |
 |
Organism: Streptococcus pyogenes, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2022-10-26 Classification: HYDROLASE Ligands: MG, K, EDO |
 |
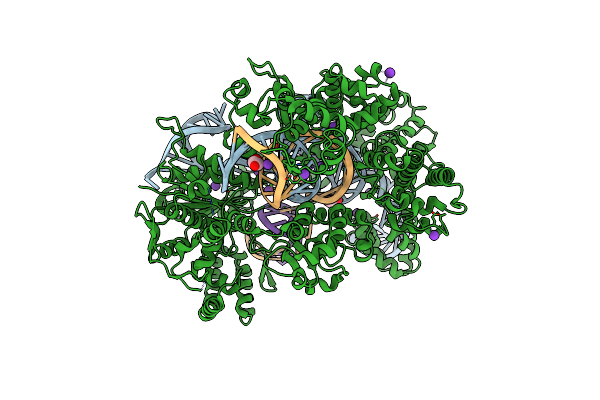
Organism: Streptococcus pyogenes, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2022-10-26 Classification: HYDROLASE Ligands: MG, K, EDO |
 |
Organism: Streptococcus pyogenes, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.75 Å Release Date: 2022-10-26 Classification: HYDROLASE Ligands: MG, K, EDO |
 |
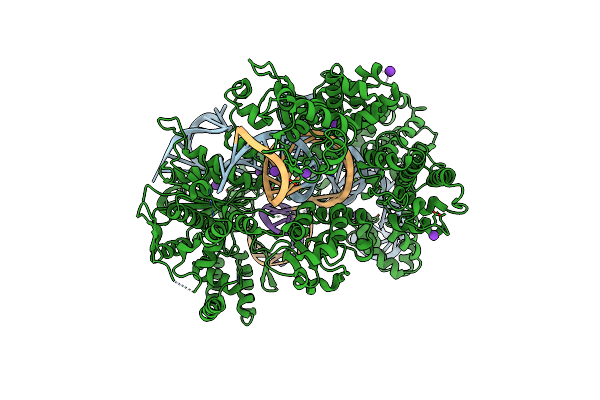
Organism: Streptococcus pyogenes, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2022-10-26 Classification: HYDROLASE Ligands: MG, K |
 |
Organism: Streptococcus pyogenes, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2022-10-26 Classification: HYDROLASE Ligands: MG, K |
 |
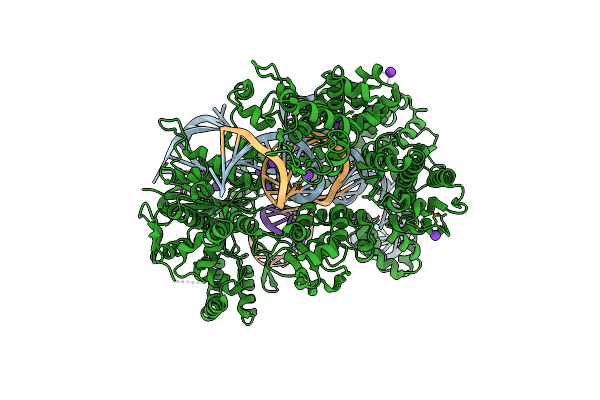
Organism: Streptococcus pyogenes, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2022-10-26 Classification: HYDROLASE Ligands: MG, K |
 |
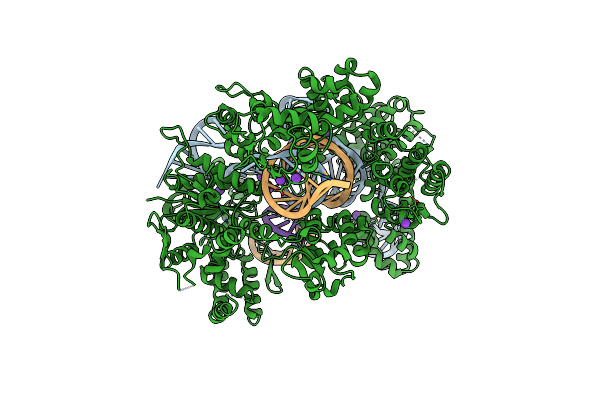
Organism: Streptococcus pyogenes, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2022-10-26 Classification: HYDROLASE Ligands: MG, K |
 |
Organism: Streptococcus pyogenes, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2022-10-26 Classification: HYDROLASE Ligands: MG, K |
 |
Organism: Streptococcus pyogenes, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2022-10-26 Classification: HYDROLASE Ligands: MG, K |

