Search Count: 73
 |
Crystal Structure Of The Thermotoga Maritima Cold Shock Protein Mutant Est#13 In Complex With Aacest
Organism: Alicyclobacillus acidocaldarius, Thermotoga maritima
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2025-01-15 Classification: PROTEIN BINDING Ligands: PMS, PEG, GOL |
 |
Organism: Geobacillus kaustophilus (strain hta426), Alicyclobacillus sp. tp-7, Alicyclobacillus acidocaldarius
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2021-09-08 Classification: ISOMERASE Ligands: MPD, MN, RB0 |
 |
Organism: Geobacillus kaustophilus (strain hta426), Alicyclobacillus sp. tp-7, Alicyclobacillus acidocaldarius
Method: X-RAY DIFFRACTION Resolution:2.49 Å Release Date: 2021-09-01 Classification: ISOMERASE Ligands: MPD, MN, GOL |
 |
Organism: Alicyclobacillus acidocaldarius subsp. acidocaldarius
Method: X-RAY DIFFRACTION Resolution:2.33 Å Release Date: 2019-02-06 Classification: OXIDOREDUCTASE Ligands: FAD |
 |
Organism: Alicyclobacillus acidocaldarius subsp. acidocaldarius (strain atcc 27009 / dsm 446 / jcm 5260 / nbrc 15652 / ncimb 11725 / nrrl b-14509 / 104-1a)
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2017-07-05 Classification: OXIDOREDUCTASE |
 |
Crystal Structure Of Icir Transcriptional Regulator From Alicyclobacillus Acidocaldarius
Organism: Alicyclobacillus acidocaldarius subsp. acidocaldarius (strain atcc 27009 / dsm 446 / jcm 5260 / nbrc 15652 / ncimb 11725 / nrrl b-14509 / 104-1a)
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2016-10-26 Classification: TRANSCRIPTION Ligands: GOL |
 |
Crystal Structure Of Single Mutant Thermostable Endoglucanase (D468A) From Alicyclobacillus Acidocaldarius
Organism: Alicyclobacillus acidocaldarius
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2016-02-03 Classification: HYDROLASE Ligands: CA, ZN, MPD |
 |
Crystal Structure Of Putative M16-Like Peptidase From Alicyclobacillus Acidocaldarius
Organism: Alicyclobacillus acidocaldarius subsp. acidocaldarius dsm 446
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2015-03-18 Classification: HYDROLASE |
 |
Organism: Alicyclobacillus acidocaldarius subsp. acidocaldarius
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2014-07-09 Classification: OXIDOREDUCTASE,ISOMERASE Ligands: MG, TLA, NDP |
 |
Crystal Structure Of Aminoglycoside Phosphotransferase From Alicyclobacillus Acidocaldarius Subsp. Acidocaldarius Dsm 446
Organism: Alicyclobacillus acidocaldarius subsp. acidocaldarius
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2014-05-07 Classification: TRANSFERASE Ligands: HIS, SCN |
 |
The Structure Of Gh113 Beta-Mannanase Aamana From Alicyclobacillus Acidocaldarius In Complex With Manifg
Organism: Alicyclobacillus acidocaldarius
Method: X-RAY DIFFRACTION Resolution:1.64 Å Release Date: 2014-04-02 Classification: HYDROLASE Ligands: IFM, BMA |
 |
The Structure Of Gh113 Beta-Mannanase Aamana From Alicyclobacillus Acidocaldarius In Complex With Manifg And Beta-1,4-Mannobiose
Organism: Alicyclobacillus acidocaldarius
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2014-04-02 Classification: HYDROLASE Ligands: IFM, BMA |
 |
The Structure Of Gh113 Beta-Mannanase Aamana From Alicyclobacillus Acidocaldarius In Complex With Manmim
Organism: Alicyclobacillus acidocaldarius
Method: X-RAY DIFFRACTION Resolution:1.47 Å Release Date: 2014-04-02 Classification: HYDROLASE Ligands: MVL, BMA |
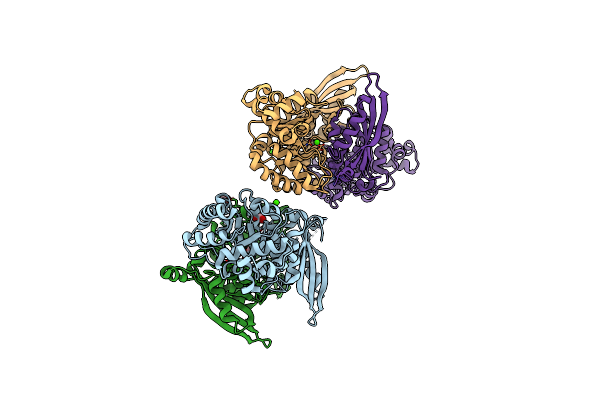 |
The Crystal Structure Of A Rubisco-Like Protein (Mtnw) From Alicyclobacillus Acidocaldarius Subsp. Acidocaldarius Dsm 446
Organism: Alicyclobacillus acidocaldarius subsp. acidocaldarius
Method: X-RAY DIFFRACTION Resolution:1.92 Å Release Date: 2013-11-13 Classification: LYASE Ligands: GOL, FMT, CA, CL |
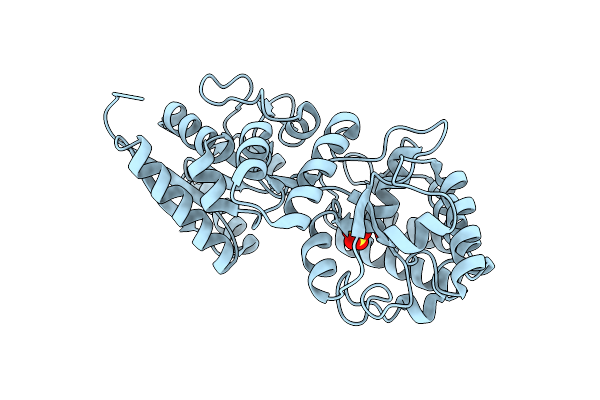 |
Extracellular Solute-Binding Protein Family 1 From Alicyclobacillus Acidocaldarius Subsp. Acidocaldarius Dsm 446
Organism: Alicyclobacillus acidocaldarius subsp. acidocaldarius (strain atcc 27009 / dsm 446 / jcm 5260 / nbrc 15652 / ncimb 11725 / nrrl b-14509 / 104-1a)
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2013-11-06 Classification: TRANSPORT PROTEIN Ligands: SO4 |
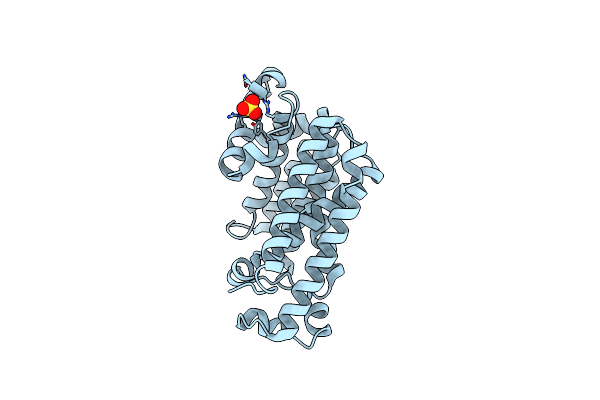 |
Crystal Structure Of Squalene Synthase Hpnc From Alicyclobacillus Acidocaldarius
Organism: Alicyclobacillus acidocaldarius subsp. acidocaldarius
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2012-10-24 Classification: TRANSFERASE Ligands: SO4 |
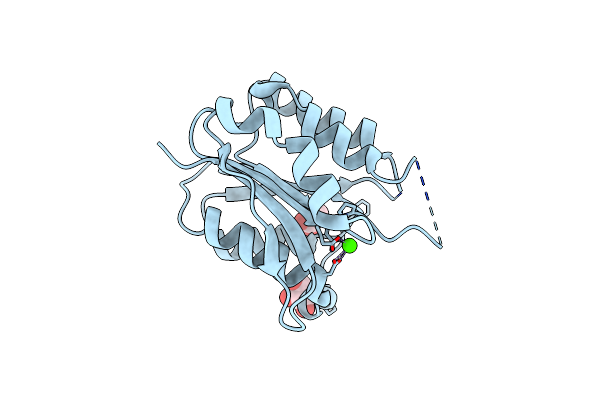 |
The Crystal Structure Of Protein-(Glutamine-N5) Methyltransferase (Release Factor-Specific) From Alicyclobacillus Acidocaldarius Subsp. Acidocaldarius Dsm 446
Organism: Alicyclobacillus acidocaldarius subsp. acidocaldarius dsm 446
Method: X-RAY DIFFRACTION Resolution:2.55 Å Release Date: 2012-03-14 Classification: TRANSFERASE Ligands: CA, ACT, GOL |
 |
The Structure Of A Xylose Isomerase Domain Protein From Alicyclobacillus Acidocaldarius Subsp. Acidocaldarius.
Organism: Alicyclobacillus acidocaldarius subsp. acidocaldarius
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2011-11-09 Classification: ISOMERASE Ligands: PO4, EDO, GOL |
 |
Crystal Structure Of Alicyclobacillus Acidocaldarius Protein With Beta-Lactamase And Rhodanese Domains
Organism: Alicyclobacillus acidocaldarius subsp. acidocaldarius
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2011-09-21 Classification: HYDROLASE Ligands: ZN |
 |
Crystal Structure Of Histidine--Trna Ligase Subunit From Alicyclobacillus Acidocaldarius Subsp. Acidocaldarius Dsm 446.
Organism: Alicyclobacillus acidocaldarius subsp. acidocaldarius
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2011-09-14 Classification: LIGASE Ligands: LMR, NA, ACY, GOL |

