Search Count: 16
 |
Cryo-Em Structure Of Microtubule-Bound Klp61F Motor Domain In The Amppnp State
Organism: Drosophila melanogaster, Sus scrofa
Method: ELECTRON MICROSCOPY Release Date: 2020-02-19 Classification: MOTOR PROTEIN Ligands: GTP, MG, GDP, ANP |
 |
Cryo-Em Structure Of Microtubule-Bound Klp61F Motor With Tail Domain In The Nucleotide-Free State
Organism: Drosophila melanogaster, Sus scrofa
Method: ELECTRON MICROSCOPY Release Date: 2020-02-19 Classification: MOTOR PROTEIN Ligands: GTP, MG, GDP |
 |
Structure Of Arabidopsis Thaliana Glr3.3 Ligand-Binding Domain In Complex With L-Glutamate
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2020-01-01 Classification: MEMBRANE PROTEIN Ligands: GLU, SO4, NA, EDO |
 |
Structure Of Arabidopsis Thaliana Glr3.3 Ligand-Binding Domain In Complex With Glycine
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2020-01-01 Classification: MEMBRANE PROTEIN Ligands: GLY, GOL, NA, CL, SO4 |
 |
Structure Of Arabidopsis Thaliana Glr3.3 Ligand-Binding Domain In Complex With L-Cysteine
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2020-01-01 Classification: MEMBRANE PROTEIN Ligands: CYS, NA, GOL, CL |
 |
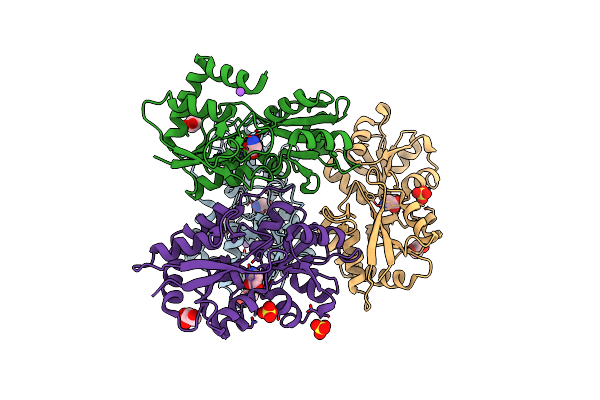
Structure Of Arabidopsis Thaliana Glr3.3 Ligand-Binding Domain In Complex With L-Methionine
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2020-01-01 Classification: MEMBRANE PROTEIN Ligands: MET, NA, SO4 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:4.31 Å Release Date: 2019-05-29 Classification: CELL CYCLE |
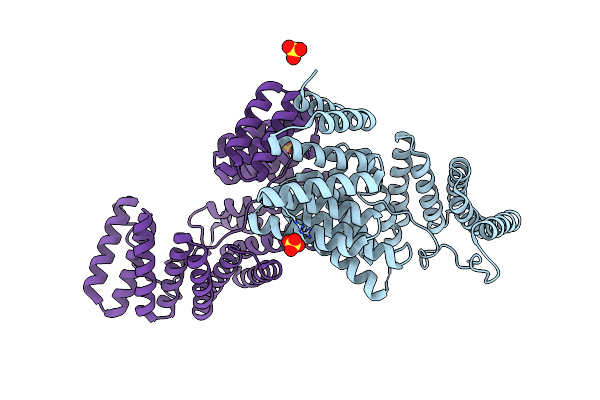 |
Concomitant Binding Of Afadin To Lgn And F-Actin Directs Planar Spindle Orientation
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2015-12-30 Classification: CELL ADHESION Ligands: SO4 |
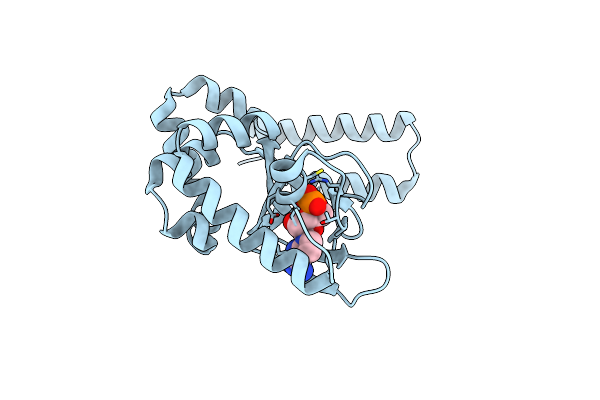 |
Structure Of The Cyclic Nucleotide-Binding Domain Of Hcn4 Channel Complexed With 7-Ch-Camp
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2014-03-19 Classification: TRANSPORT PROTEIN Ligands: 7CH |
 |
Organism: Drosophila melanogaster
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2012-03-28 Classification: CELL CYCLE |
 |
Bacterial Flavin-Containing Monooxygenase In Complex With Nadp: Native Data
Organism: Methylophaga sp. sk1
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2008-04-22 Classification: OXIDOREDUCTASE Ligands: FAD, NAP, CL, EPE, PGE, PG4 |
 |
Bacterial Flavin-Containing Monooxygenase In Complex With Nadp: Soaking In Aerated Solution
Organism: Methylophaga sp. sk1
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2008-04-22 Classification: OXIDOREDUCTASE Ligands: FAD, NAP, CL, EPE, OXY |
 |
Structure Of The Monooxygenase Component Of P-Hydroxyphenylacetate Hydroxylase From Acinetobacter Baumanni
Organism: Acinetobacter baumannii
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2007-01-23 Classification: OXIDOREDUCTASE |
 |
Structure Of The Monooxygenase Component Of P-Hydroxyphenylacetate Hydroxylase From Acinetobacter Baumannii
Organism: Acinetobacter baumannii
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2007-01-23 Classification: OXIDOREDUCTASE Ligands: FMN |
 |
Structure Of The Monooxygenase Component Of P-Hydroxyphenylacetate Hydroxylase From Acinetobacter Baumannii
Organism: Acinetobacter baumannii
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2007-01-23 Classification: OXIDOREDUCTASE Ligands: FMN, 4HP |
 |
Organism: Thermobifida fusca
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2004-09-02 Classification: OXYGENASE Ligands: FAD, SO4 |

