Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
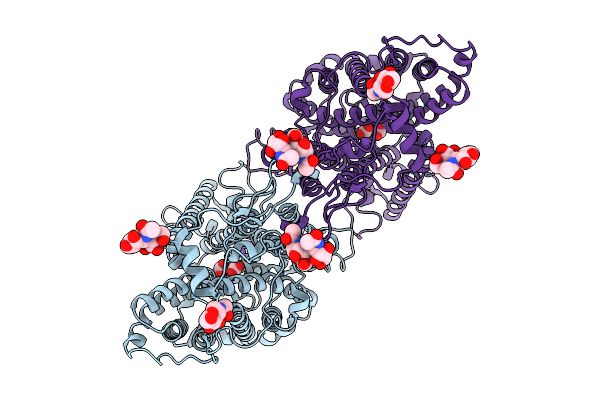 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-09-03 Classification: PROTEIN TRANSPORT Ligands: CO3, NAG, NA |
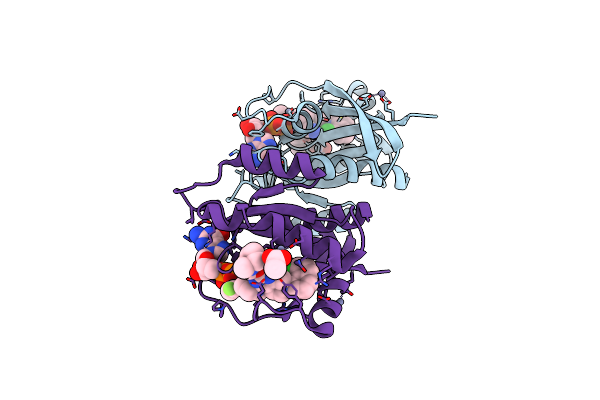 |
Crystal Structure Of Wild-Type Kras (Gdp-Bound) In Complex With Mrtx849 (Adagrasib)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-06-25 Classification: ONCOPROTEIN/INHIBITOR Ligands: GDP, ZN, A1B7W, ACT |
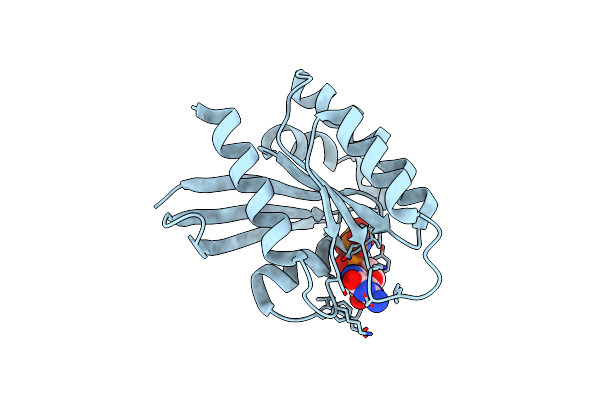 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-06-25 Classification: ONCOPROTEIN Ligands: GNP, MG |
 |
Crystal Structure Of Active Kras-G12C (Gmppnp-Bound) In Complex With Bbo-8520
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2024-12-18 Classification: ONCOPROTEIN/INHIBITOR Ligands: GNP, MG, Y8N |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.67 Å Release Date: 2024-12-18 Classification: ONCOPROTEIN/INHIBITOR Ligands: GDP, MG, Y8N |
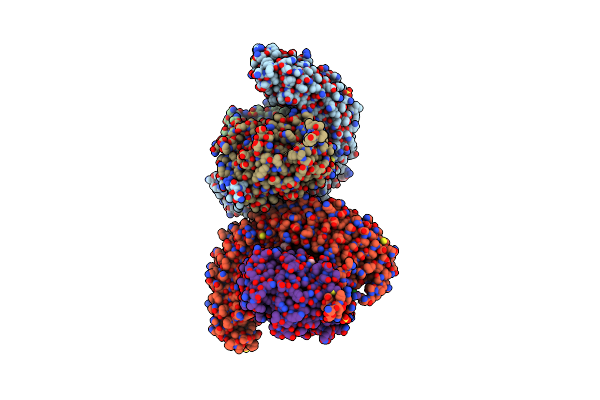 |
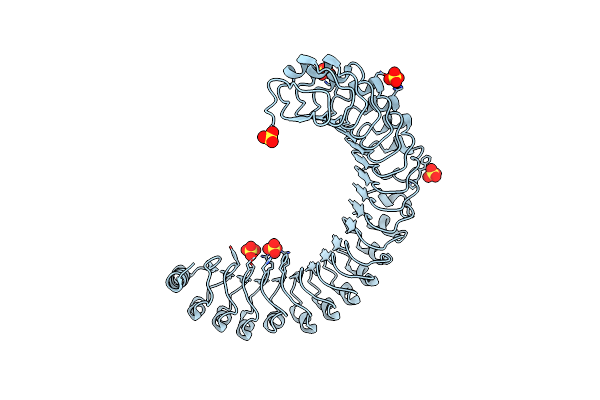
Crystal Structure Of The Shoc2-Mras-Pp1Ca (Smp) Complex To A Resolution Of 2.17 Angstrom
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.17 Å Release Date: 2022-05-04 Classification: SIGNALING PROTEIN Ligands: GOL, SO4, MN, NA, PO4, CL, GNP, MG |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2022-05-04 Classification: SIGNALING PROTEIN Ligands: SO4, CL |
 |
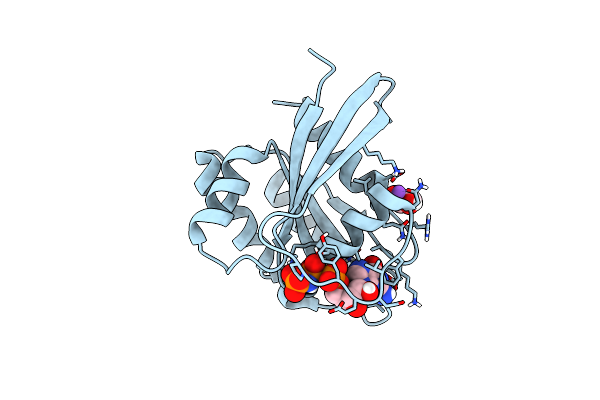
Structure Of Mg-Free Kras4B (2-169) Bound To Gdp With The Switch-I In Fully Open Conformation
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2019-07-31 Classification: ONCOPROTEIN Ligands: GDP |
 |
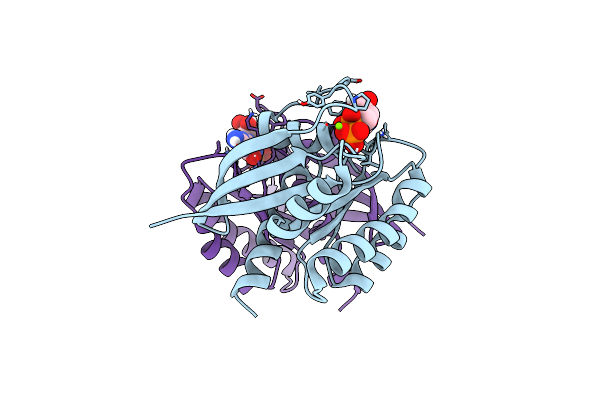
Crystal Structure Of Mg-Free Wild-Type Kras (2-166) Bound To Gmppnp In The State 1 Conformation
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2019-07-31 Classification: ONCOPROTEIN |
 |
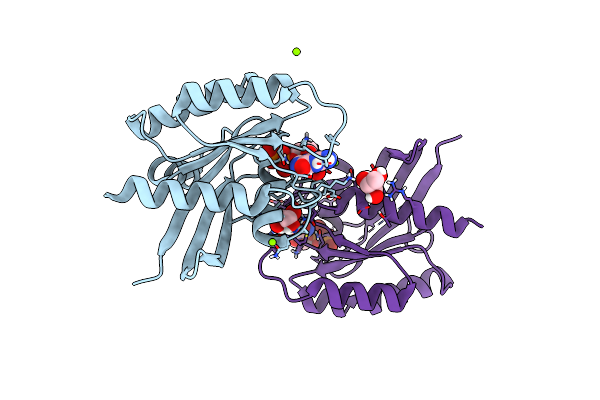
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2019-07-31 Classification: ONCOPROTEIN Ligands: GDP, MG |
 |
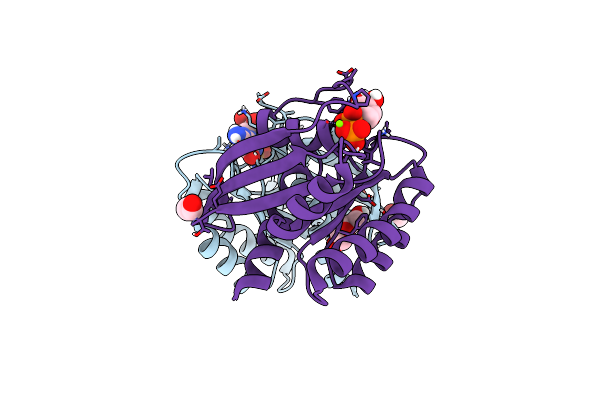
Crystal Structure Of Wild-Type Kras (1-169) Bound To Gdp And Mg (Space Group P3)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2019-07-31 Classification: ONCOPROTEIN Ligands: MG, GDP, GOL |
 |
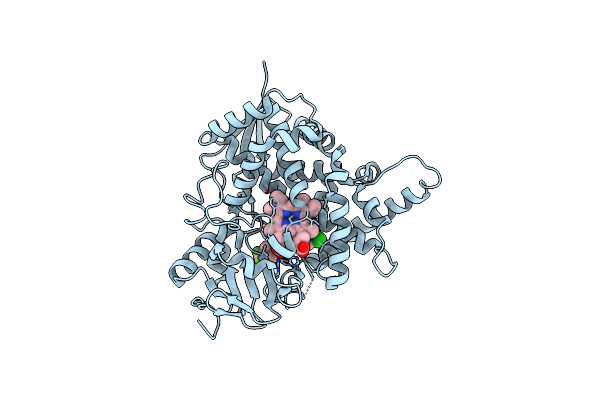
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.01 Å Release Date: 2019-07-31 Classification: ONCOPROTEIN Ligands: MG, GDP, ACE, PEG |
 |
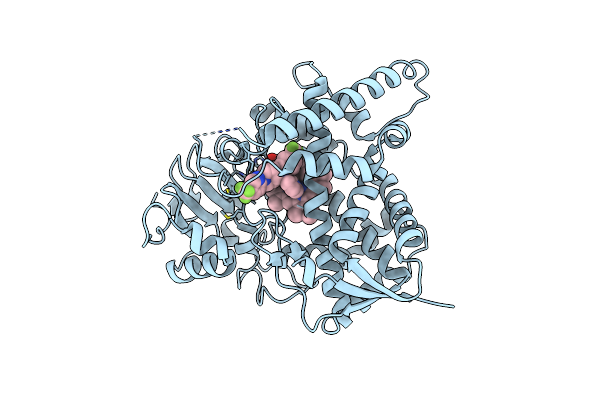
Sterol 14 Alpha-Demethylase (Cyp51) From Trypanosoma Cruzi In Complex With The Pyridine Inhibitor (S)-2-(4-Chlorophenyl)-2-(Pyridin-3-Yl)-1- (4-(4-(Trifluoromethyl)Phenyl)Piperazin-1-Yl)Ethanone (Epl-Bs1246,Udo)
Organism: Trypanosoma cruzi
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2013-09-25 Classification: OXIDOREDUCTASE Ligands: HEM, UDO |
 |
Sterol 14-Alpha Demethylase (Cyp51)From Trypanosoma Cruzi In Complex With The Pyridine Inhibitor N-(1-(5-(Trifluoromethyl)(Pyridin-2-Yl)) Piperidin-4Yl)-N-(4-(Trifluoromethyl)Phenyl)Pyridin-3-Amine (Epl- Bs967, Udd)
Organism: Trypanosoma cruzi
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2013-09-25 Classification: OXIDOREDUCTASE Ligands: HEM, UDD |
 |
Organism: Artificial gene
Method: SOLUTION NMR Release Date: 2012-02-29 Classification: DE NOVO PROTEIN |
 |
Organism: Artificial gene
Method: SOLUTION NMR Release Date: 2012-02-29 Classification: DE NOVO PROTEIN |
 |
Organism: Artificial gene
Method: SOLUTION NMR Release Date: 2012-02-29 Classification: DE NOVO PROTEIN |
 |
Organism: Artificial gene
Method: SOLUTION NMR Release Date: 2012-02-29 Classification: DE NOVO PROTEIN |
 |
Nmr Structures Of Ga95 And Gb95, Two Designed Proteins With 95% Sequence Identity But Different Folds And Functions
|
 |
Nmr Structures Of Ga95 And Gb95, Two Designed Proteins With 95% Sequence Identity But Different Folds And Functions
|