Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
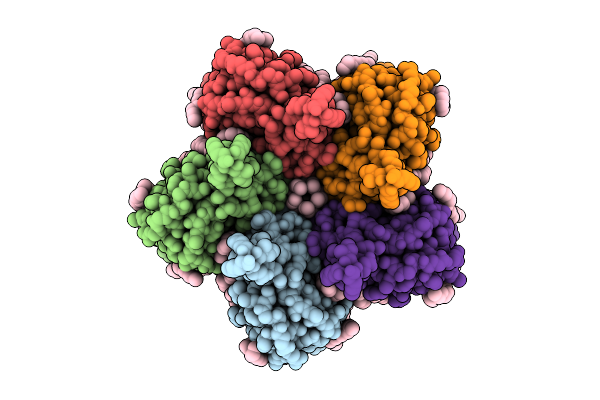 |
Cryo-Em Structure Of The Flotillin-Associated Rhodopsin Psfar In Detergent Micelle
Organism: Candidatus pseudothioglobus
Method: ELECTRON MICROSCOPY Release Date: 2025-07-23 Classification: MEMBRANE PROTEIN Ligands: LFA |
 |
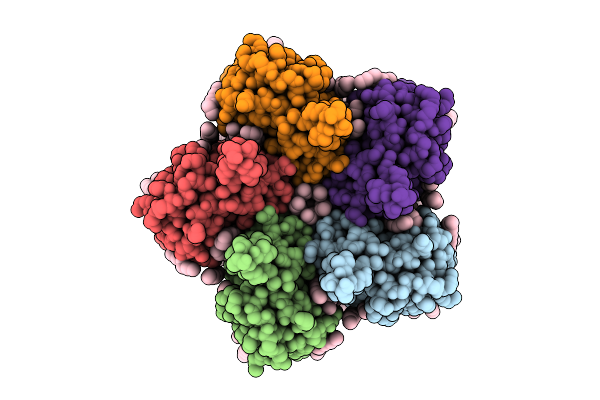
Cryo-Em Structure Of The Light-Driven Proton Pump Pspr In Detergent Micelle
Organism: Candidatus pseudothioglobus sp.
Method: ELECTRON MICROSCOPY Release Date: 2025-07-23 Classification: MEMBRANE PROTEIN Ligands: LFA, RET, LMT |
 |
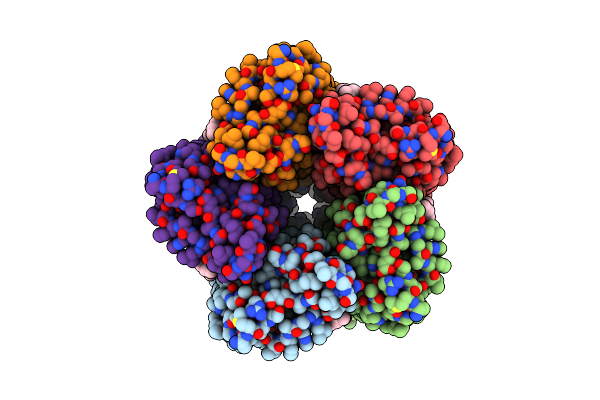
Cryo-Em Structure Of The Double Mutant H84V/E120G Of The Flotillin-Associated Rhodopsin Psfar In Detergent Micelle
Organism: Candidatus pseudothioglobus sp.
Method: ELECTRON MICROSCOPY Release Date: 2025-07-23 Classification: MEMBRANE PROTEIN Ligands: LFA, RET |
 |
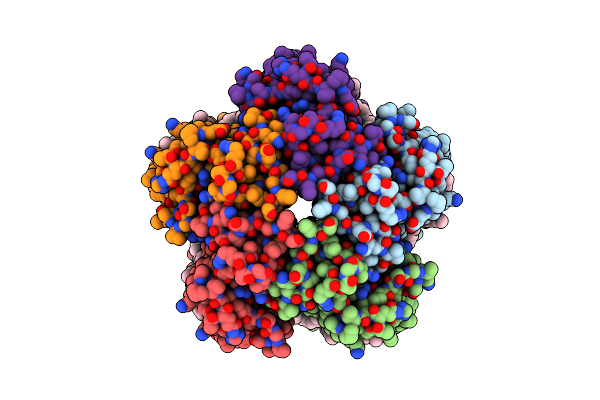
Organism: Cryobacterium levicorallinum
Method: ELECTRON MICROSCOPY Release Date: 2025-05-14 Classification: MEMBRANE PROTEIN Ligands: LFA, RET |
 |
Organism: Cryobacterium levicorallinum
Method: ELECTRON MICROSCOPY Release Date: 2025-05-14 Classification: MEMBRANE PROTEIN Ligands: LFA, RET |
 |
Organism: Cryobacterium levicorallinum
Method: ELECTRON MICROSCOPY Release Date: 2025-05-14 Classification: MEMBRANE PROTEIN Ligands: LMT, LFA, RET |
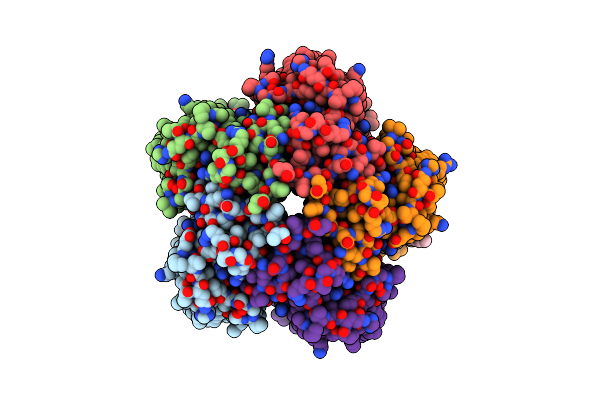 |
Cryo-Em Structure Of The Microbial Rhodopsin Cryor1 At Ph 10.5 In Detergent In The Ground State
Organism: Cryobacterium levicorallinum
Method: ELECTRON MICROSCOPY Release Date: 2025-05-14 Classification: MEMBRANE PROTEIN Ligands: LMT, RET, LFA |
 |
Cryo-Em Structure Of The Microbial Rhodopsin Cryor1 At Ph 10.5 In Detergent In The M State
Organism: Cryobacterium levicorallinum
Method: ELECTRON MICROSCOPY Release Date: 2025-05-14 Classification: MEMBRANE PROTEIN Ligands: LMT, RET |
 |
Organism: Subtercola endophyticus
Method: ELECTRON MICROSCOPY Release Date: 2025-05-14 Classification: MEMBRANE PROTEIN Ligands: LFA, RET |
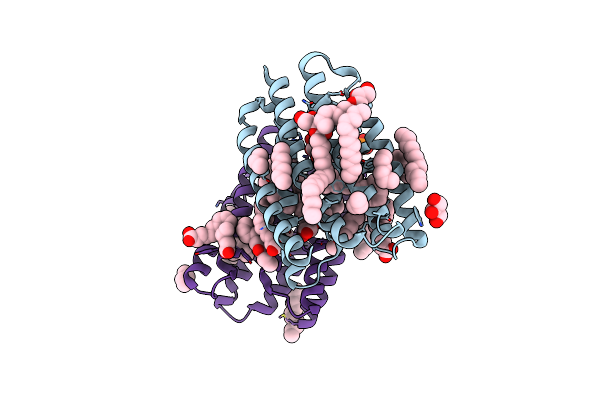 |
Crystal Structure Of Marine Actinobacteria Clade Rhodopsin (Mar) In The Ground State
Organism: Candidatus actinomarina minuta, Marine actinobacteria clade
Method: X-RAY DIFFRACTION Release Date: 2025-04-02 Classification: MEMBRANE PROTEIN Ligands: OLC, LFA, GOL, RET, PO4 |
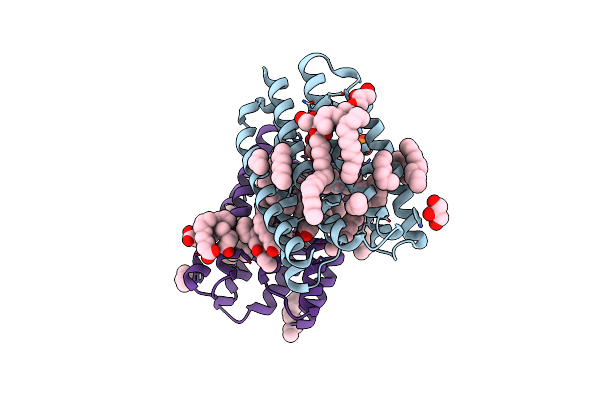 |
Crystal Structure Of Marine Actinobacteria Clade Rhodopsin (Mar) In The P596 State
Organism: Candidatus actinomarina minuta, Marine actinobacteria clade
Method: X-RAY DIFFRACTION Release Date: 2025-04-02 Classification: MEMBRANE PROTEIN Ligands: OLC, LFA, GOL, RET, PO4 |
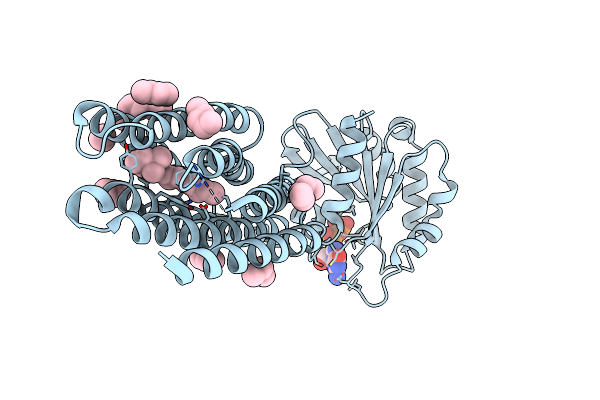 |
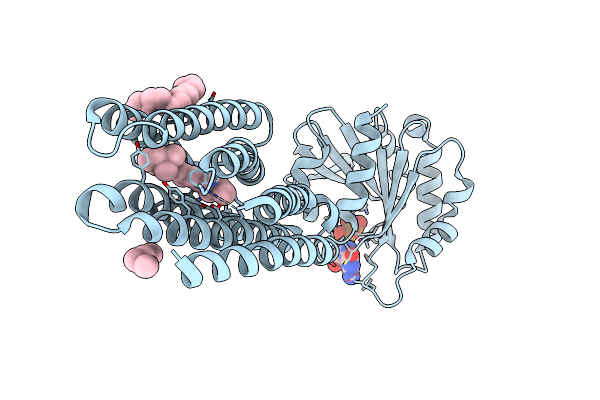
Crystal Structure Of Marine Actinobacteria Clade Rhodopsin (Mar) - Human Gtpase Arf1 (L8K,Q71L) Chimera; Ground State
Organism: Candidatus actinomarina minuta, Homo sapiens, Marine actinobacteria clade
Method: X-RAY DIFFRACTION Release Date: 2025-04-02 Classification: MEMBRANE PROTEIN Ligands: GDP, LFA, RET |
 |
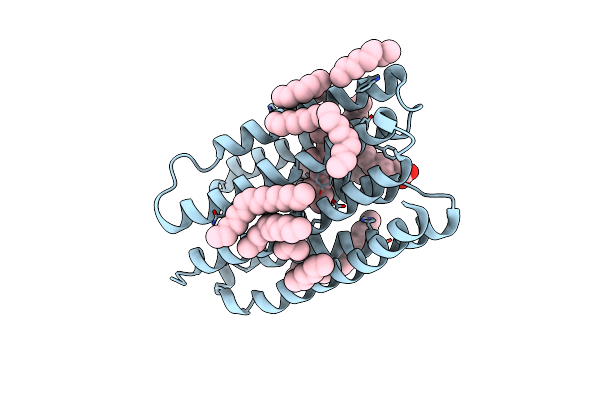
Crystal Structure Of Marine Actinobacteria Clade Rhodopsin (Mar) - Human Gtpase Arf1 (L8K,Q71L) Chimera; N State
Organism: Candidatus actinomarina minuta, Homo sapiens, Marine actinobacteria clade
Method: X-RAY DIFFRACTION Release Date: 2025-04-02 Classification: MEMBRANE PROTEIN Ligands: GDP, LFA, OLA, RET |
 |
Crystal Structure Of Marine Actinobacteria Clade Rhodopsin (Mar) In The O* State
Organism: Candidatus actinomarina minuta, Marine actinobacteria clade
Method: X-RAY DIFFRACTION Release Date: 2025-04-02 Classification: MEMBRANE PROTEIN Ligands: OLA, LFA, RET |
 |
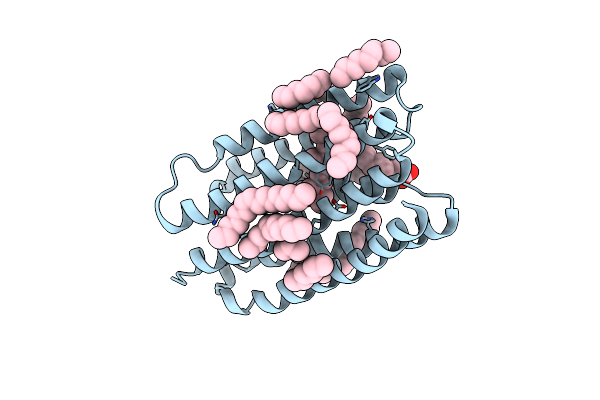
Crystal Structure Of Marine Actinobacteria Clade Rhodopsin (Mar) In The O* State, Ph 8.8
Organism: Candidatus actinomarina minuta, Marine actinobacteria clade
Method: X-RAY DIFFRACTION Release Date: 2025-04-02 Classification: MEMBRANE PROTEIN Ligands: OLA, LFA, RET |
 |
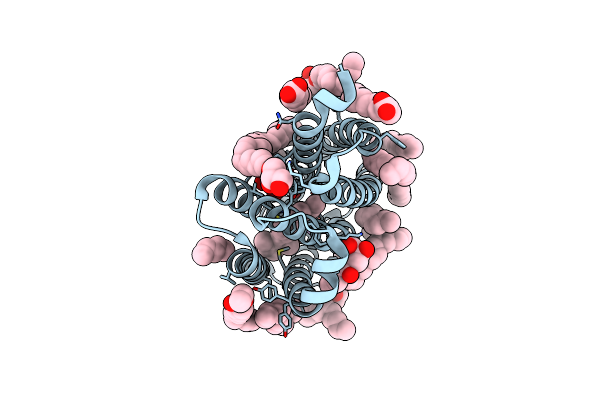
Crystal Structure Of Marine Actinobacteria Clade Rhodopsin (Mar) In The O State Obtained By Cryotrapping
Organism: Candidatus actinomarina minuta, Marine actinobacteria clade
Method: X-RAY DIFFRACTION Release Date: 2025-04-02 Classification: MEMBRANE PROTEIN Ligands: OLA, LFA, RET |
 |
True-Atomic Resolution Crystal Structure Of The Closed State Of The Viral Channelrhodopsin Olpvr1
Organism: Organic lake phycodnavirus
Method: X-RAY DIFFRACTION Resolution:1.13 Å Release Date: 2025-03-12 Classification: MEMBRANE PROTEIN |
 |
True-Atomic Resolution Crystal Structure Of The Open State Of The Viral Channelrhodopsin Olpvr1
Organism: Organic lake phycodnavirus
Method: X-RAY DIFFRACTION Resolution:1.34 Å Release Date: 2025-03-12 Classification: MEMBRANE PROTEIN Ligands: RET, NA, 97N, OLC, LFA, GOL |
 |
Structure Of The Viral Channelrhodopsin Olpvr1 At Low Ph Obtained By Soaking
Organism: Organic lake phycodnavirus
Method: X-RAY DIFFRACTION Resolution:1.41 Å Release Date: 2025-03-12 Classification: MEMBRANE PROTEIN Ligands: RET, 97N, LFA, OLC |
 |
Crystal Structure Of The Light-Driven Sodium Pump Ernar In The Monomeric Form At Ph 4.6
Organism: Erythrobacter
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2024-04-24 Classification: MEMBRANE PROTEIN Ligands: LFA, OLA |