Search Count: 142
All
Selected
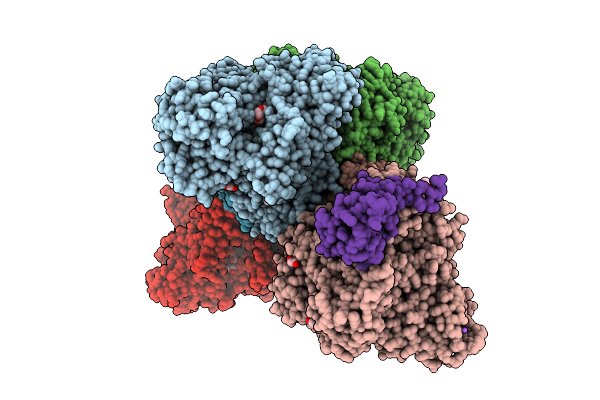 |
Organism: Alcaligenes faecalis
Method: X-RAY DIFFRACTION Resolution:1.89 Å Release Date: 2025-08-06 Classification: OXIDOREDUCTASE Ligands: MGD, 4MO, F3S, PEG, EDO, 1PE, PGE, GOL, O, NA, FES, P4G, PG0 |
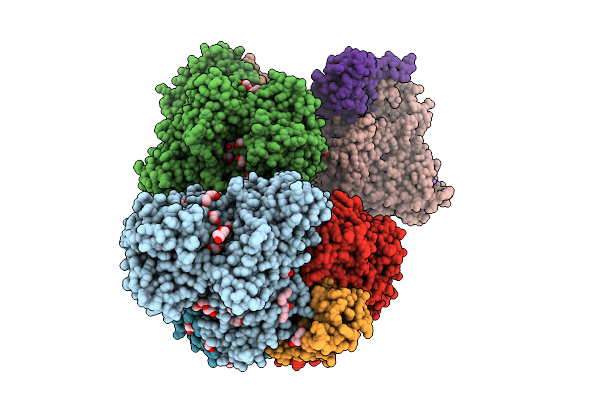 |
Organism: Alcaligenes faecalis
Method: X-RAY DIFFRACTION Resolution:1.87 Å Release Date: 2025-08-06 Classification: OXIDOREDUCTASE Ligands: MO, MGD, F3S, EDO, PEG, SBO, P33, PGE, O, NA, FES, GOL, 4MO, P4G, 1PE, PG0 |
 |
Organism: Alcaligenes faecalis
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2025-02-26 Classification: METAL BINDING PROTEIN Ligands: ZN |
 |
Crystal Structure Of Pyruvic Oxime Dioxygenase (Pod) From Alcaligenes Faecalis Deleted N-Terminal 18 Residues
Organism: Alcaligenes faecalis subsp. faecalis nbrc 13111
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2024-03-20 Classification: OXIDOREDUCTASE Ligands: NI |
 |
Crystal Structure Of Pyruvic Oxime Dioxygenase (Pod) From Alcaligenes Faecalis
Organism: Alcaligenes faecalis subsp. faecalis nbrc 13111
Method: X-RAY DIFFRACTION Resolution:1.77 Å Release Date: 2024-03-06 Classification: OXIDOREDUCTASE Ligands: FE2 |
 |
Crystal Structure Of Arsenite Oxidase From Alcaligenes Faecalis (Af Aio) Bound To Arsenic Oxyanion
Organism: Alcaligenes faecalis
Method: X-RAY DIFFRACTION Resolution:1.43 Å Release Date: 2024-02-21 Classification: OXIDOREDUCTASE Ligands: MGD, F3S, FC6, 4MO, ART, GOL, EDO, FES, PEG |
 |
Organism: Alcaligenes faecalis
Method: X-RAY DIFFRACTION Resolution:0.86 Å Release Date: 2024-02-14 Classification: ELECTRON TRANSPORT Ligands: CU, SO4 |
 |
Organism: Alcaligenes faecalis
Method: X-RAY DIFFRACTION, NEUTRON DIFFRACTION Resolution:1.50 Å, 1.90 Å Release Date: 2024-02-14 Classification: ELECTRON TRANSPORT Ligands: CU, SO4, DOD |
 |
Crystal Structure Of Arsenite Oxidase From Alcaligenes Faecalis (Af Aio) Bound To Arsenite
Organism: Alcaligenes faecalis
Method: X-RAY DIFFRACTION Resolution:1.57 Å Release Date: 2023-07-12 Classification: OXIDOREDUCTASE Ligands: MGD, MO, F3S, PGE, GOL, AST, EDO, PEG, IPA, FES |
 |
Crystal Structure Of Arsenite Oxidase From Alcaligenes Faecalis (Af Aio) Bound To Antimony Oxyanion
Organism: Alcaligenes faecalis
Method: X-RAY DIFFRACTION Resolution:1.84 Å Release Date: 2023-07-12 Classification: OXIDOREDUCTASE Ligands: MGD, O, 4MO, F3S, PEG, ACT, PO4, UJI, CL, GOL, TRS, FES, EDO, SO4, NA |
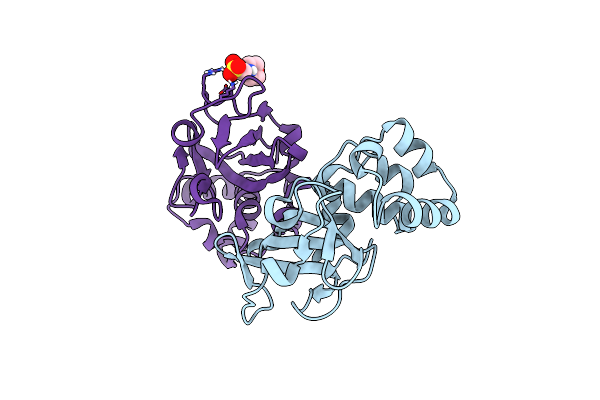 |
Structure Of The Bacterial Toxin, Teca, An Asparagine Deamidase From Alcaligenes Faecalis.
Organism: Alcaligenes faecalis
Method: X-RAY DIFFRACTION Resolution:1.84 Å Release Date: 2022-11-02 Classification: TOXIN Ligands: MES |
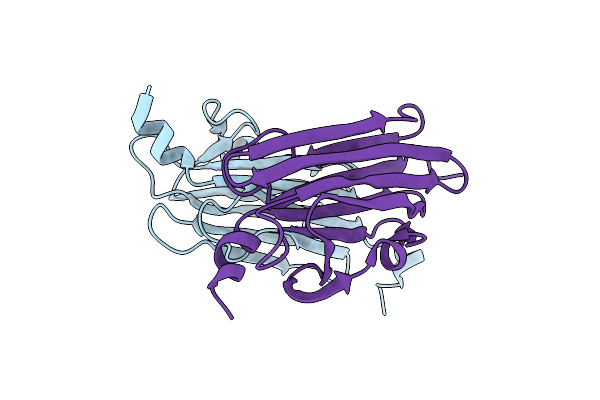 |
Organism: Alcaligenes faecalis
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2017-06-14 Classification: TOXIN |
 |
Organism: Alcaligenes faecalis
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2017-04-12 Classification: ELECTRON TRANSPORT Ligands: CU |
 |
Crystal Structure Of D-3-Hydroxybutyrate Dehydrogenase From Alcaligenes Faecalis Complexed With Nad+ And A Substrate D-3-Hydroxybutyrate
Organism: Alcaligenes faecalis
Method: X-RAY DIFFRACTION Resolution:1.19 Å Release Date: 2016-08-17 Classification: OXIDOREDUCTASE Ligands: NAD, 3HR, NA |
 |
Crystal Structure Of D-3-Hydroxybutyrate Dehydrogenase From Alcaligenes Faecalis Complexed With Nad+ And An Inhibitor Malonate
Organism: Alcaligenes faecalis
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2016-08-17 Classification: OXIDOREDUCTASE/OXIDOREDUCTASE INHIBITOR Ligands: NAD, MLA, NA, CL |
 |
Crystal Structure Of D-3-Hydroxybutyrate Dehydrogenase From Alcaligenes Faecalis Complexed With Nad+ And An Inhibitor Methylmalonate
Organism: Alcaligenes faecalis
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2016-08-17 Classification: OXIDOREDUCTASE/OXIDOREDUCTASE INHIBITOR Ligands: NAD, DXX, CL, NA |
 |
Nitrite Complex Structure Of Copper Nitrite Reductase From Alcaligenes Faecalis Determined At 293 K
Organism: Alcaligenes faecalis
Method: X-RAY DIFFRACTION Resolution:1.54 Å Release Date: 2016-03-16 Classification: OXIDOREDUCTASE Ligands: CU, NO2 |
 |
Resting State Structure Of Cunir Form Alcaligenes Faecalis Determined At 293 K
Organism: Alcaligenes faecalis
Method: X-RAY DIFFRACTION Resolution:1.56 Å Release Date: 2016-03-16 Classification: OXIDOREDUCTASE Ligands: CU |
 |
Completely Oxidized Structure Of Copper Nitrite Reductase From Alcaligenes Faecalis
Organism: Alcaligenes faecalis
Method: X-RAY DIFFRACTION Resolution:2.03 Å Release Date: 2016-03-09 Classification: OXIDOREDUCTASE Ligands: CU, CL |
 |
High Resolution Synchrotron Structure Of Copper Nitrite Reductase From Alcaligenes Faecalis
Organism: Alcaligenes faecalis
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2016-03-09 Classification: OXIDOREDUCTASE Ligands: CU, MPD, ACY |

