Search Count: 6
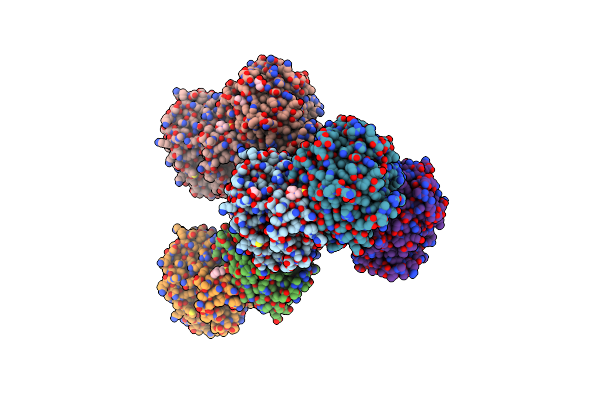 |
Crystal Structure Of Mgs-Mt1, An Alpha/Beta Hydrolase Enzyme From A Lake Matapan Deep-Sea Metagenome Library
Organism: Unidentified
Method: X-RAY DIFFRACTION Resolution:1.74 Å Release Date: 2015-03-04 Classification: HYDROLASE Ligands: CL, MES, GOL |
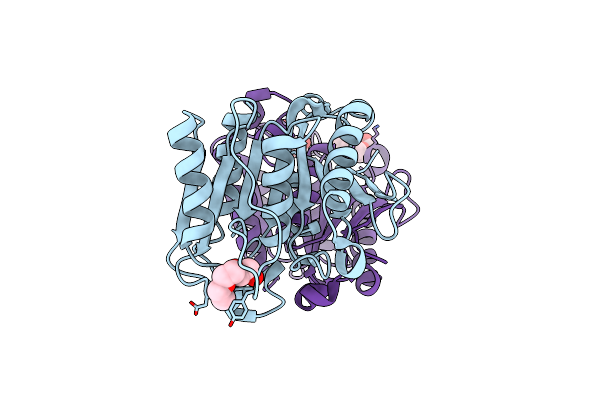 |
Crystal Structure Of Mgs-M1, An Alpha/Beta Hydrolase Enzyme From A Medee Basin Deep-Sea Metagenome Library
Organism: Unidentified
Method: X-RAY DIFFRACTION Resolution:1.57 Å Release Date: 2015-02-25 Classification: HYDROLASE Ligands: CL, F, PGE |
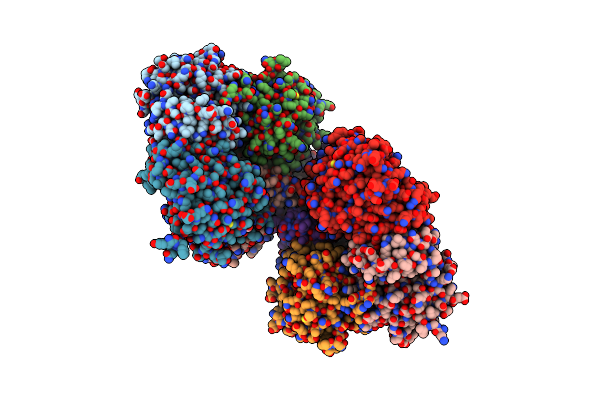 |
Crystal Structure Of Mgs-M2, An Alpha/Beta Hydrolase Enzyme From A Medee Basin Deep-Sea Metagenome Library
Organism: Unidentified
Method: X-RAY DIFFRACTION Resolution:3.01 Å Release Date: 2015-02-25 Classification: HYDROLASE Ligands: GOL |
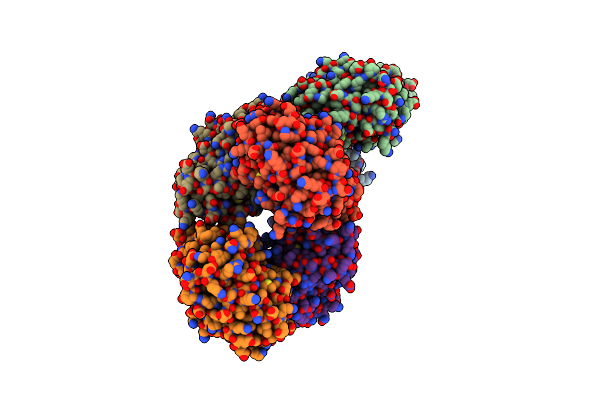 |
Crystal Structure Of Mgs-M4, An Aldo-Keto Reductase Enzyme From A Medee Basin Deep-Sea Metagenome Library
Organism: Unidentified
Method: X-RAY DIFFRACTION Resolution:2.55 Å Release Date: 2015-02-25 Classification: HYDROLASE Ligands: SO4, NA |
 |
Crystal Structure Of Mgs-M5, A Lactate Dehydrogenase Enzyme From A Medee Basin Deep-Sea Metagenome Library
Organism: Unidentified
Method: X-RAY DIFFRACTION Resolution:1.97 Å Release Date: 2015-02-25 Classification: HYDROLASE |
 |
Crystal Structure Of Serine Hydrolase Ccsp0084 From The Polyaromatic Hydrocarbon (Pah)-Degrading Bacterium Cycloclasticus Zankles
Organism: Cycloclasticus sp. p1
Method: X-RAY DIFFRACTION Resolution:1.69 Å Release Date: 2013-06-26 Classification: HYDROLASE Ligands: CL, NA, PEG |

