Search Count: 15
 |
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2024-12-11 Classification: TRANSFERASE |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.30 Å Release Date: 2024-12-11 Classification: TRANSFERASE |
 |
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2023-04-19 Classification: TRANSFERASE/INHIBITOR Ligands: WIB, EDO |
 |
Identification Of A Class Of Wnk Isoform-Specific Inhibitors Through High-Throughput Screening
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.11 Å Release Date: 2023-03-01 Classification: TRANSFERASE Ligands: WGK |
 |
Crystallography Of Novel Wnk1 And Wnk3 Inhibitors Discovered From High-Throughput-Screening
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2020-04-15 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: MU7, GOL, ACT |
 |
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Release Date: 2018-06-20 Classification: TRANSFERASE Ligands: ANP |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.84 Å Release Date: 2014-05-28 Classification: TRANSFERASE Ligands: PO4 |
 |
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:3.50 Å Release Date: 2014-05-28 Classification: TRANSFERASE Ligands: BR |
 |
The Third Conformation Of P38A Map Kinase Observed In Phosphorylated P38A And In Solution
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2011-01-05 Classification: TRANSFERASE |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2009-03-03 Classification: TRANSFERASE Ligands: SO4, GOL, EDO |
 |
The X-Ray Crystal Structure Of The Paramecium Bursaria Chlorella Virus Arginine Decarboxylase
Organism: Paramecium bursaria chlorella virus 1
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2007-03-20 Classification: LYASE Ligands: PLP |
 |
The X-Ray Crystal Structure Of The Paramecium Bursaria Chlorella Virus Arginine Decarboxylase Bound To Agmatine
Organism: Paramecium bursaria chlorella virus 1
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2007-03-20 Classification: LYASE Ligands: PL2 |
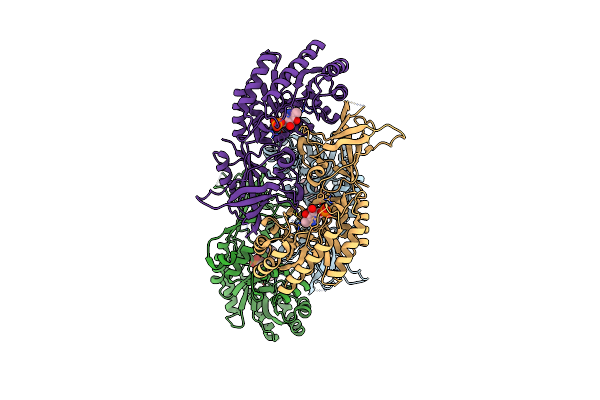 |
A Dimer Interface Mutant Of Ornithine Decarboxylase Reveals Structure Of Gem Diamine Intermediate
Organism: Trypanosoma brucei brucei
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2004-10-26 Classification: LYASE Ligands: PXP, PLG, ORX |
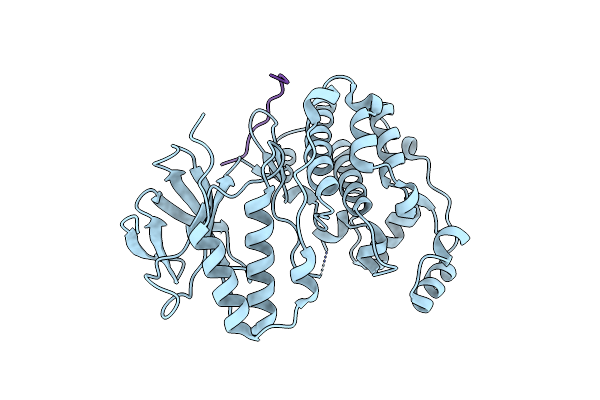 |
Crystal Structure Of Map Kinase P38 Complexed To The Docking Site On Its Nuclear Substrate Mef2A
Organism: Mus musculus, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2002-07-10 Classification: TRANSFERASE |
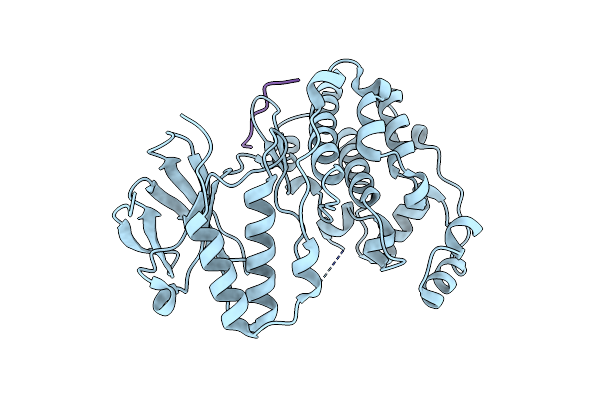 |
Crystal Structure Of Map Kinase P38 Complexed To The Docking Site On Its Activator Mkk3B
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2002-07-10 Classification: TRANSFERASE |

