Search Count: 49
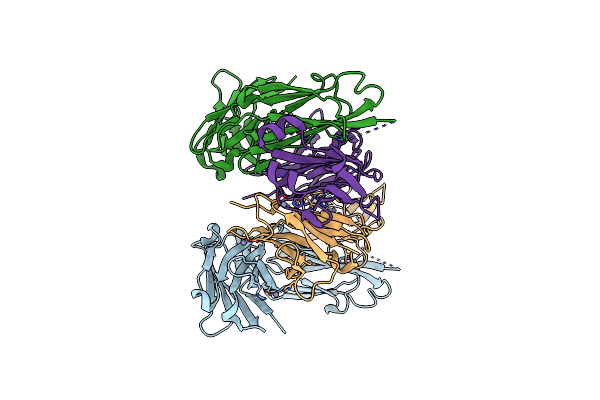 |
Crystal Structure Of Fab From Mouse Monoclonal Antibody 4A9 Against Aquifex Aeolicus Rsep Pdz Tandem
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2025-03-05 Classification: PROTEIN BINDING Ligands: ZN |
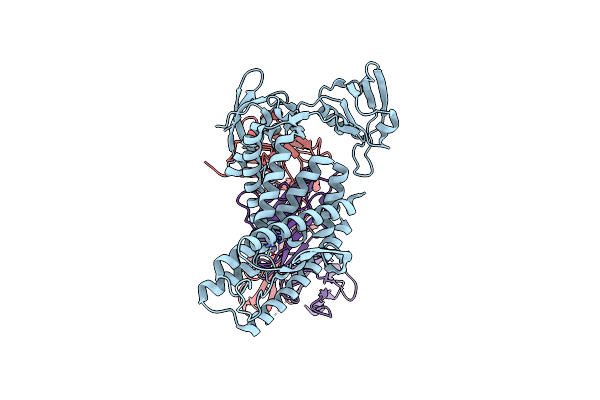 |
Organism: Aquifex aeolicus vf5, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2025-03-05 Classification: HYDROLASE Ligands: ZN |
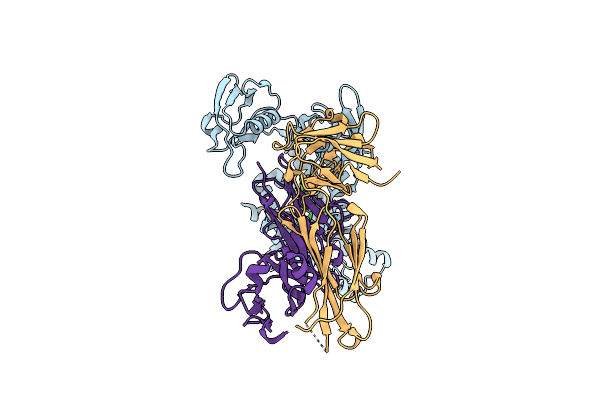 |
Organism: Aquifex aeolicus vf5, Mus musculus, Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2025-03-05 Classification: HYDROLASE Ligands: ZN |
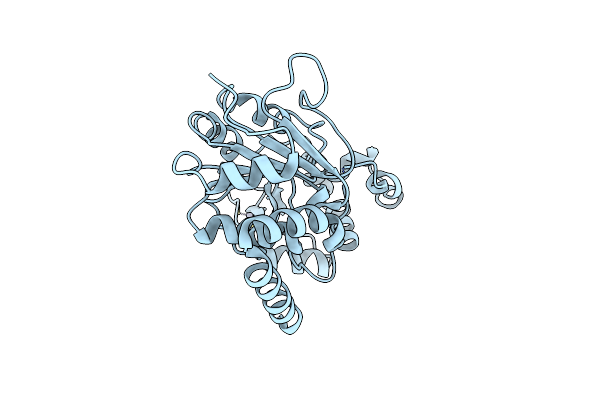 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2024-05-22 Classification: METAL BINDING PROTEIN Ligands: ZN |
 |
The Structural Model Of A Homodimeric D-Ala-D-Ala Metallopeptidase, Vanx, From Vancomycin-Resistant Bacteria
Organism: Enterococcus faecium
Method: SOLUTION NMR Release Date: 2024-05-22 Classification: METAL BINDING PROTEIN |
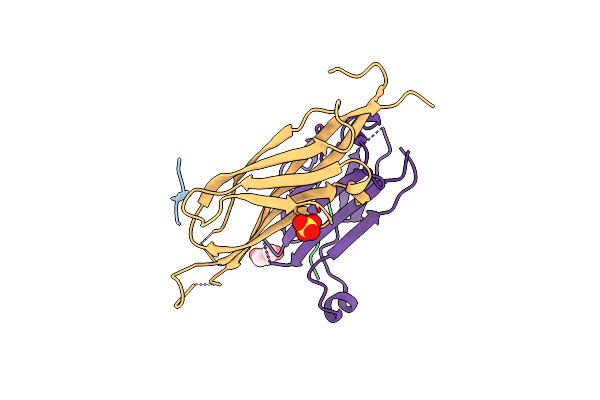 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2023-07-26 Classification: PROTEIN BINDING Ligands: SO4, GOL |
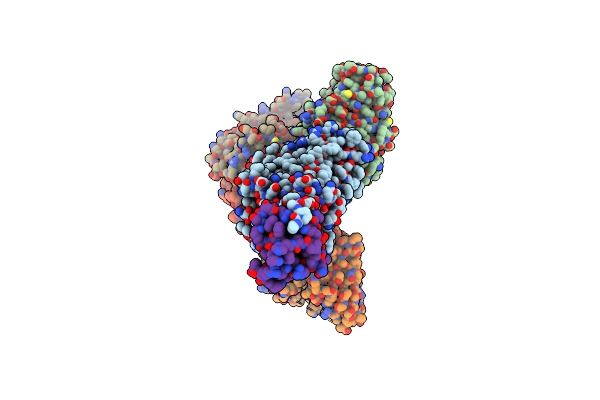 |
Cryo-Em Structure Of Cxcl8 Bound C-X-C Chemokine Receptor 1 In Complex With Gi Heterotrimer
Organism: Homo sapiens, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2023-07-19 Classification: MEMBRANE PROTEIN/IMMUNE SYSTEM |
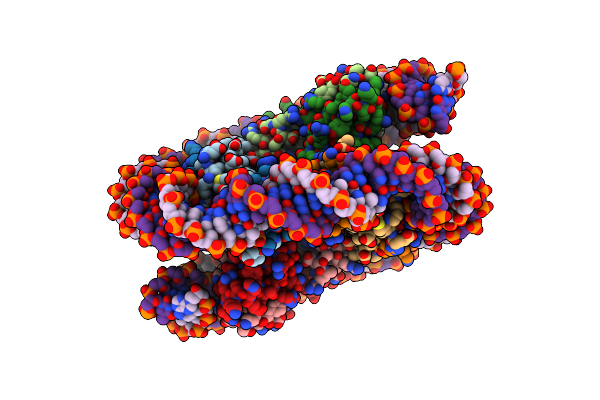 |
Organism: Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2022-11-16 Classification: DNA BINDING PROTEIN/DNA |
 |
Organism: Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2022-11-16 Classification: DNA BINDING PROTEIN/DNA |
 |
Organism: Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2022-11-16 Classification: DNA BINDING PROTEIN/DNA |
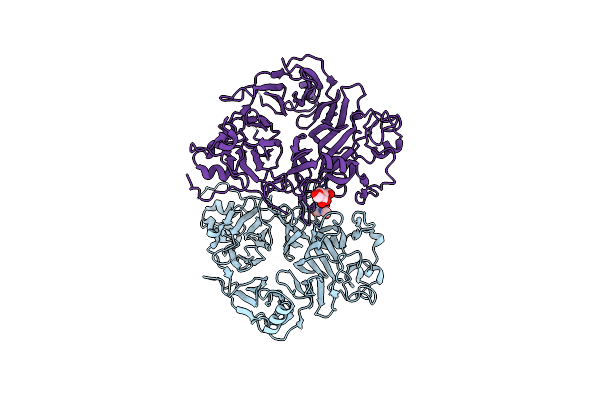 |
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2022-10-19 Classification: SIGNALING PROTEIN Ligands: NAG |
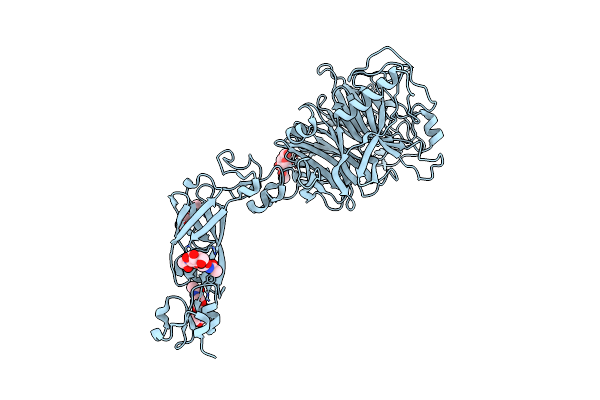 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.50 Å Release Date: 2022-10-19 Classification: SIGNALING PROTEIN Ligands: NAG |
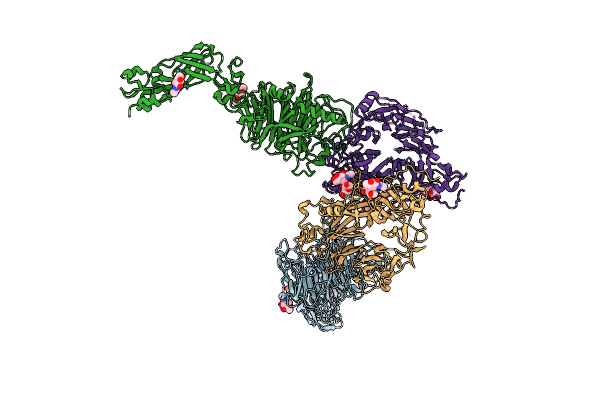 |
Organism: Homo sapiens, Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:4.70 Å Release Date: 2022-10-19 Classification: SIGNALING PROTEIN Ligands: NAG |
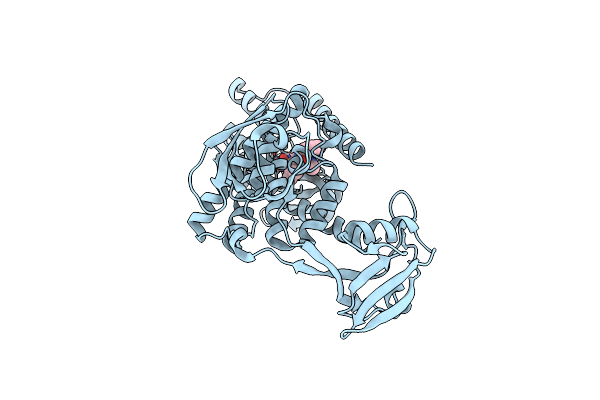 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2022-09-07 Classification: HYDROLASE Ligands: ZN, BAT |
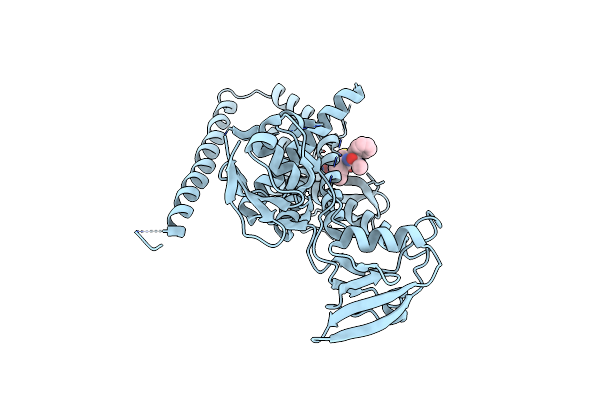 |
Crystal Structure Of Kangiella Koreensis Rsep Orthologue In Complex With Batimastat In Space Group P1
Organism: Kangiella koreensis dsm 16069
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2022-09-07 Classification: HYDROLASE Ligands: ZN, BAT |
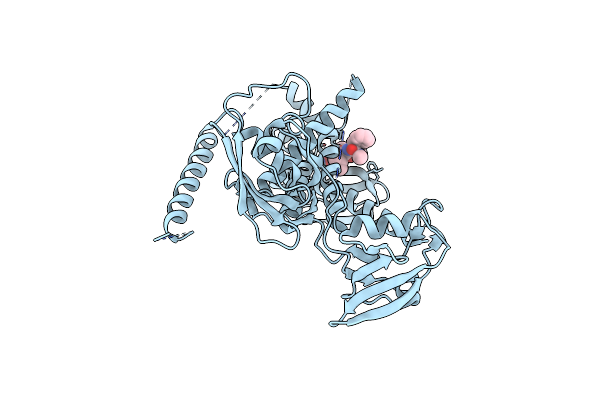 |
Crystal Structure Of Kangiella Koreensis Rsep Orthologue In Complex With Batimastat In Space Group P21
Organism: Kangiella koreensis dsm 16069
Method: X-RAY DIFFRACTION Resolution:3.15 Å Release Date: 2022-09-07 Classification: HYDROLASE Ligands: ZN, BAT |
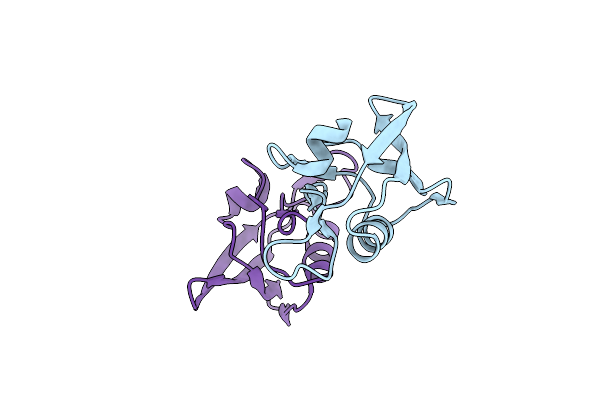 |
Crystal Structure Of The Pdz-C Domain Fragment Of Kangiella Koreensis Rsep Orthologue
Organism: Kangiella koreensis dsm 16069
Method: X-RAY DIFFRACTION Resolution:1.15 Å Release Date: 2022-09-07 Classification: HYDROLASE |
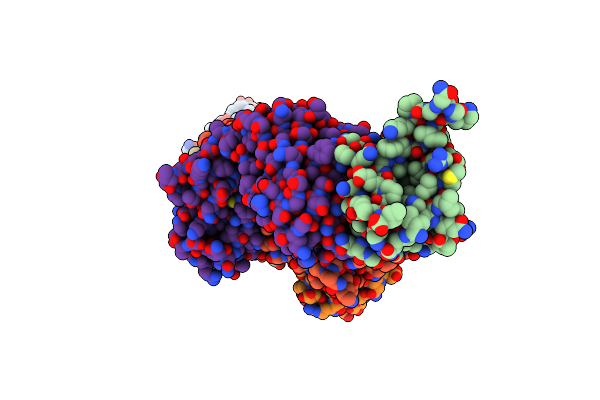 |
Crystal Structure Of The Pdz-C Domain Of E. Coli Rsep In Complex With 12C7 Fab
Organism: Escherichia coli, Mus musculus
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2022-09-07 Classification: HYDROLASE/IMMUNE SYSTEM |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2020-05-06 Classification: DE NOVO PROTEIN Ligands: GOL |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2020-05-06 Classification: DE NOVO PROTEIN Ligands: PO4, GOL |

