Search Count: 19
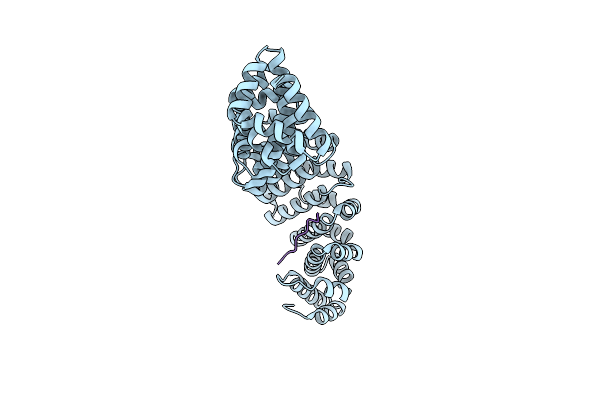 |
Crystal Structure Of Mouse Importin-Alpha1 Bound To The Nuclear Localization Signal Of Epstein-Barr Virus Ebna-Lp Protein
Organism: Mus musculus, Human herpesvirus 4 (strain b95-8)
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2017-04-19 Classification: PROTEIN TRANSPORT/TRANSCRIPTION |
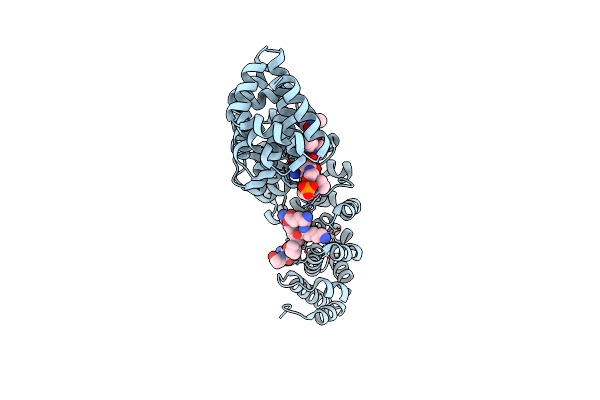 |
Crystal Structure Of Mouse Importin-Alpha1 Bound To S385-Phosphorylated Nls Of Ebna1
Organism: Mus musculus, Epstein-barr virus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2017-01-25 Classification: PROTEIN TRANSPORT/PEPTIDE |
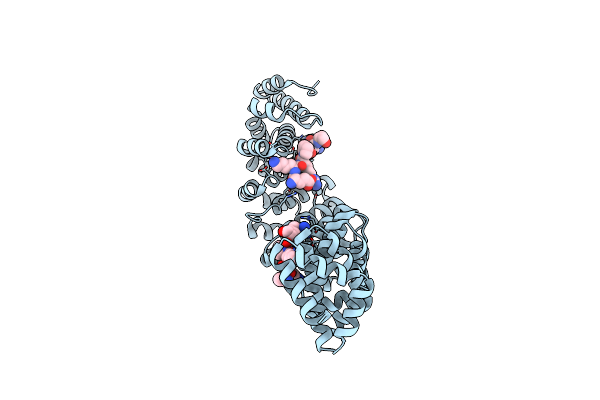 |
Crystal Structure Of Mouse Importin-Alpha1 Bound To Non-Phosphorylated Nls Of Ebna1
Organism: Mus musculus, Epstein-barr virus
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2017-01-25 Classification: PROTEIN TRANSPORT/PEPTIDE |
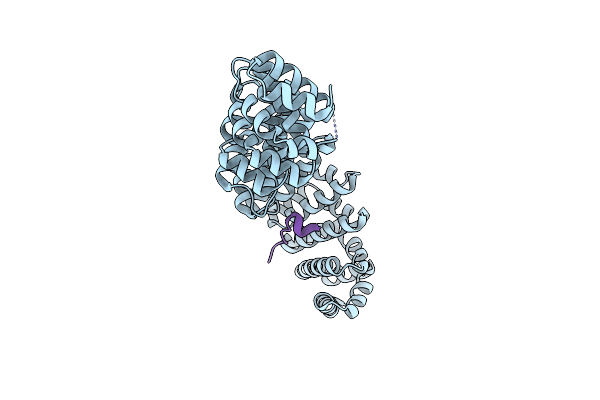 |
Crystal Structure Of Importin-Alpha Bound To A Non-Classical Nuclear Localization Signal Of The Influenza A Virus Nucleoprotein
Organism: Mus musculus, Influenza a virus (a/puerto rico/8/1934(h1n1))
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2015-10-21 Classification: PROTEIN TRANSPORT/SIGNALING PROTEIN |
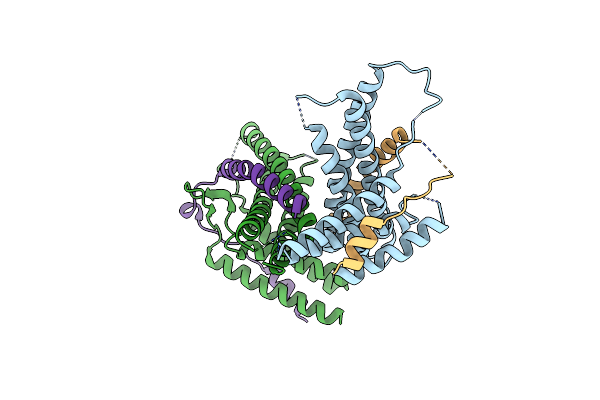 |
Organism: Kluyveromyces marxianus
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2014-05-07 Classification: PROTEIN TRANSPORT |
 |
Organism: Lachancea thermotolerans
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2014-05-07 Classification: PROTEIN TRANSPORT |
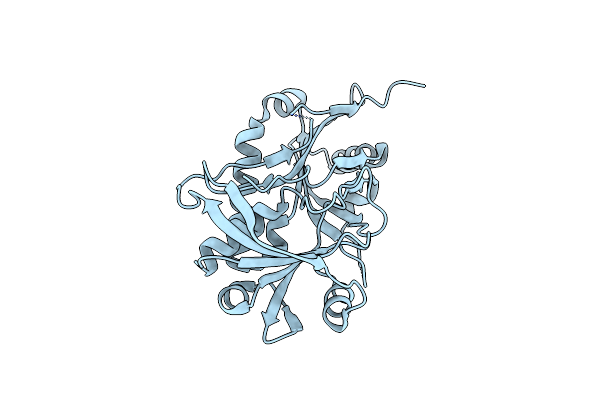 |
Organism: Kluyveromyces marxianus
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2012-11-14 Classification: LIGASE |
 |
Organism: Kluyveromyces marxianus
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2012-11-14 Classification: LIGASE |
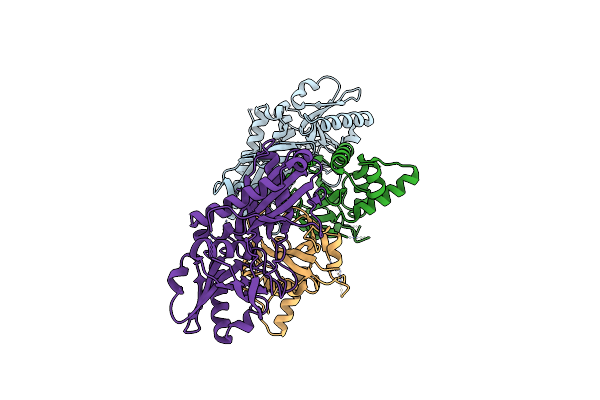 |
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:3.11 Å Release Date: 2012-11-14 Classification: LIGASE |
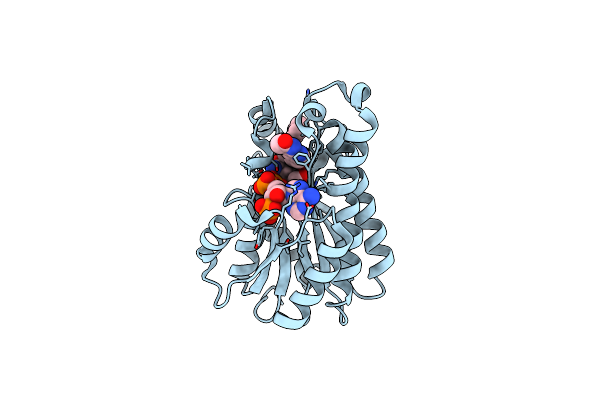 |
Crystal Structure Of Staphylococcus Aureus Enoyl-Acp Reductase In Complex With Nadp And Afn-1252
Organism: Staphylococcus aureus subsp. aureus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2012-09-19 Classification: OXIDOREDUCTASE Ligands: 0WD, 0WE |
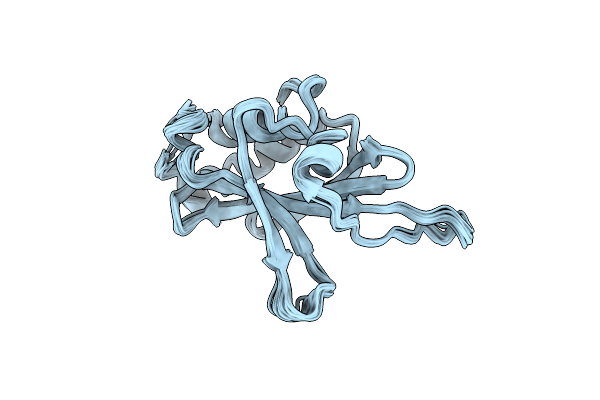 |
Organism: Kluyveromyces marxianus
Method: SOLUTION NMR Release Date: 2012-08-01 Classification: PROTEIN TRANSPORT |
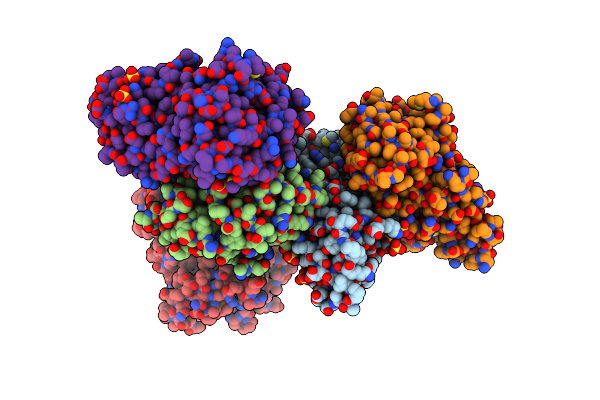 |
Organism: Kluyveromyces marxianus
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2012-08-01 Classification: PROTEIN TRANSPORT Ligands: EPE, SO4 |
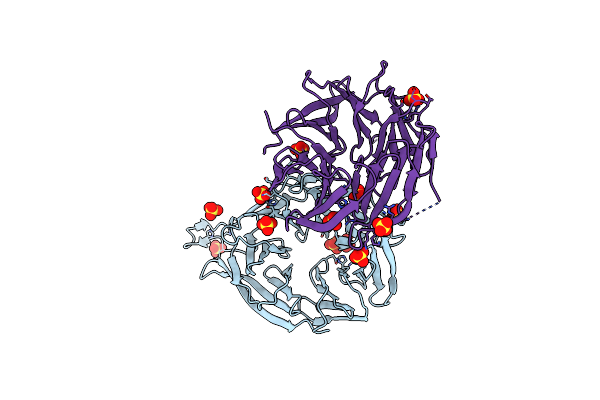 |
Organism: Kluyveromyces marxianus
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2012-07-04 Classification: PROTEIN TRANSPORT Ligands: SO4 |
 |
Crystal Structure Of Carbohydrate-Binding Module Family 28 From Clostridium Josui Cel5A In Complex With Cellopentaose
Organism: Clostridium josui
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2010-03-31 Classification: HYDROLASE Ligands: CA, PO4 |
 |
Crystal Structure Of Carbohydrate-Binding Module Family 28 From Clostridium Josui Cel5A In A Ligand-Free Form
Organism: Clostridium josui
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2010-03-02 Classification: HYDROLASE Ligands: CA, SO4 |
 |
Crystal Structure Of Carbohydrate-Binding Module Family 28 From Clostridium Josui Cel5A In Complex With Cellobiose
Organism: Clostridium josui
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2010-03-02 Classification: HYDROLASE Ligands: CA, PO4, GOL |
 |
Crystal Structure Of Carbohydrate-Binding Module Family 28 From Clostridium Josui Cel5A In Complex With Cellotetraose
Organism: Clostridium josui
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2010-03-02 Classification: HYDROLASE Ligands: CA, PO4 |
 |
 |

