Search Count: 1,015
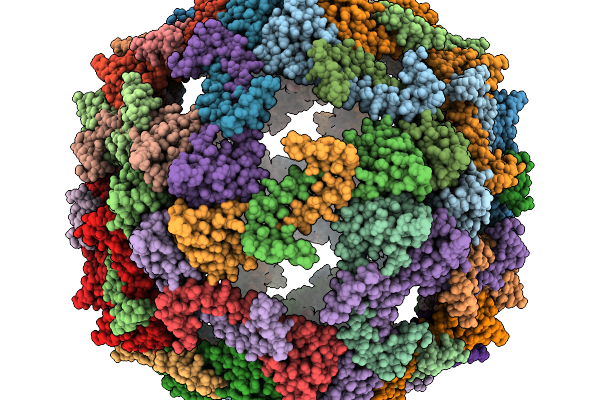 |
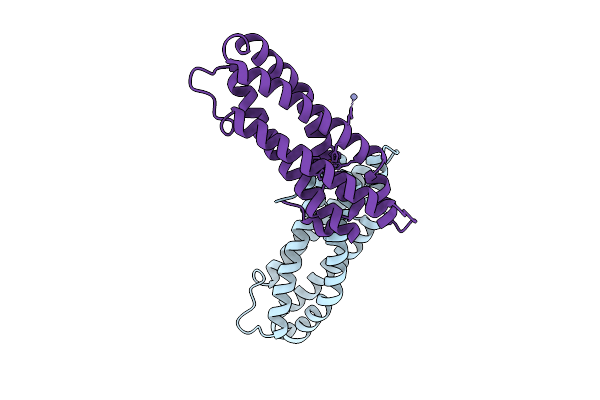
Cryo-Em Structure Of Pyrene-Modified Tip60 Double Mutant (G12C/S50C) With Addition Of Nile Red
Organism: Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: DE NOVO PROTEIN Ligands: A1L9F |
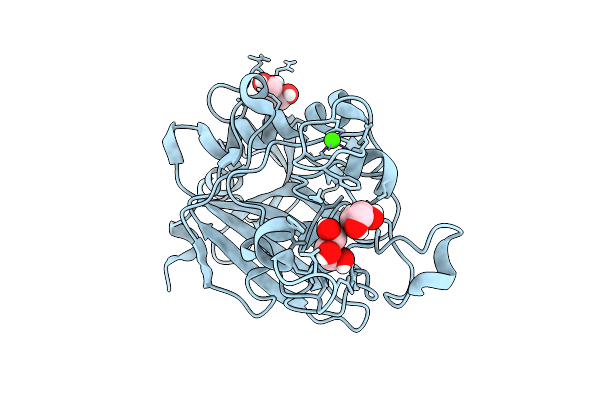 |
Crystal Structure Of Wild-Type Human Fibrinogen Gamma Chain C-Terminal Domain (Gamma-Nodule)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-11-12 Classification: BLOOD CLOTTING Ligands: GOL, CA |
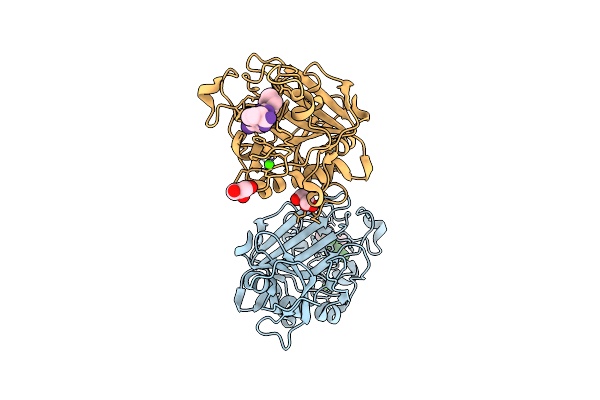 |
Crystal Structure Of Wild-Type Human Fibrinogen Gamma Chain C-Terminal Domain (Gamma-Nodule) Complexed With Gprp Peptide
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-11-12 Classification: BLOOD CLOTTING Ligands: CA, GOL |
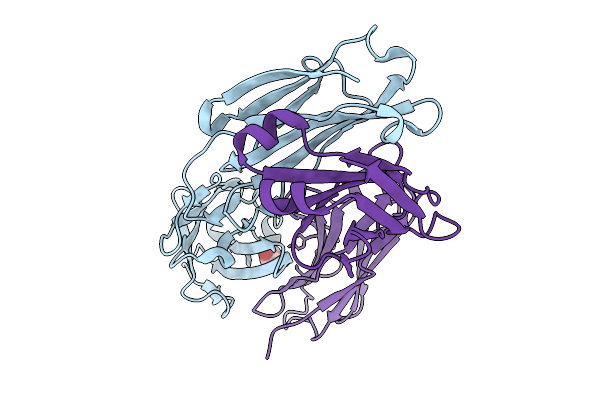 |
Crystal Structure Of Switchbody Based On Anti-Osteocalcin Antibody Ktm219 Fab
Organism: Mus musculus
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: IMMUNE SYSTEM Ligands: GOL |
 |
Organism: Squalus acanthias
Method: ELECTRON MICROSCOPY Release Date: 2025-10-01 Classification: MEMBRANE PROTEIN Ligands: CLR, PCW, MG, A1MA6 |
 |
Cryo-Em Structure Of Palytoxin-Bound Na+,K+-Atpase In The Transient State Of Dephosphorylation (E2~P)
Organism: Squalus acanthias
Method: ELECTRON MICROSCOPY Release Date: 2025-10-01 Classification: MEMBRANE PROTEIN Ligands: CLR, PCW, ALF, MG, NA, A1MA6 |
 |
Cryo-Em Structure Of Na+,K+-Atpase That Forms A Cation Channel With Palytoxin (Atp Form)
Organism: Squalus acanthias
Method: ELECTRON MICROSCOPY Release Date: 2025-10-01 Classification: MEMBRANE PROTEIN Ligands: CLR, PCW, MG, ATP, NA, A1MA6 |
 |
Cryo-Em Structure Of Na+,K+-Atpase That Forms A Cation Channel With Palytoxin (Adp Form)
Organism: Squalus acanthias
Method: ELECTRON MICROSCOPY Release Date: 2025-10-01 Classification: MEMBRANE PROTEIN Ligands: CLR, PCW, MG, ADP, NA, A1MA6 |
 |
Organism: Sus scrofa
Method: X-RAY DIFFRACTION Release Date: 2025-10-01 Classification: MEMBRANE PROTEIN Ligands: MG, CLR, PCW, A1MA6, NAG |
 |
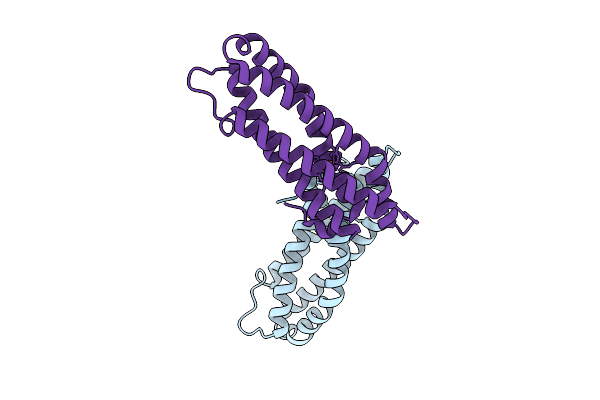
Crystal Structure Of Staphylococcus Aureus Cysteine-Free Scda With Bound Iron, Determined By Molecular Replacement And Fe Anomalous Signal
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Release Date: 2025-09-17 Classification: METAL BINDING PROTEIN Ligands: FE, O, ZN |
 |
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Release Date: 2025-08-13 Classification: OXYGEN BINDING Ligands: FE, O |
 |
Organism: Streptomyces atratus
Method: X-RAY DIFFRACTION Release Date: 2025-06-18 Classification: OXIDOREDUCTASE Ligands: HEM, CL, ACT, GOL |
 |
Organism: Streptomyces atratus
Method: X-RAY DIFFRACTION Release Date: 2025-06-18 Classification: OXIDOREDUCTASE Ligands: HEM, GOL |
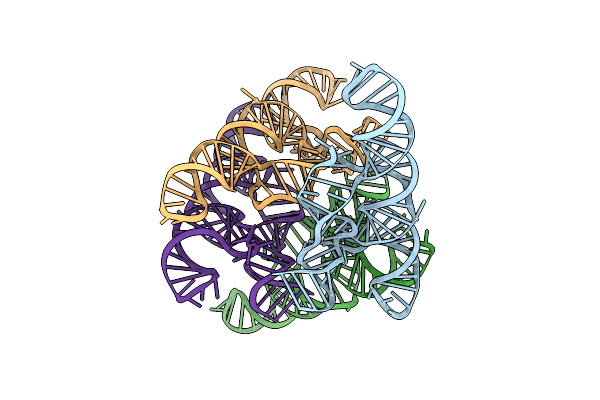 |
The Escherichia Coli Yybp Riboswitch As A Tandem Riboswitch Regulated By Mn2+ And Ph
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:3.73 Å Release Date: 2025-06-04 Classification: RNA Ligands: MG, MN |
 |
Organism: Oryza sativa japonica group
Method: X-RAY DIFFRACTION Release Date: 2025-05-21 Classification: HYDROLASE Ligands: A1EBP, GOL, BEZ, PDO, BU1, PGO |
 |
Organism: Homo sapiens, Lama glama
Method: ELECTRON MICROSCOPY Release Date: 2025-04-30 Classification: MEMBRANE PROTEIN Ligands: A1LXR |
 |
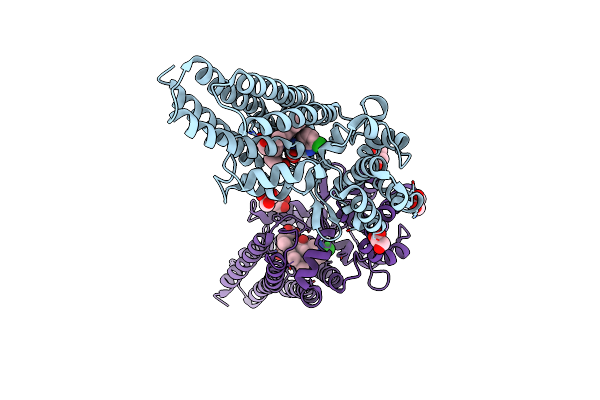
Organism: Homo sapiens, Mus musculus, Lama glama
Method: ELECTRON MICROSCOPY Release Date: 2025-04-23 Classification: MEMBRANE PROTEIN Ligands: 16C, CLR |
 |
Organism: Flavobacteriales bacterium
Method: X-RAY DIFFRACTION Resolution:2.42 Å Release Date: 2025-04-16 Classification: OXIDOREDUCTASE Ligands: GOL, HEM, CTE |
 |
Organism: Flavobacteriales bacterium
Method: X-RAY DIFFRACTION Resolution:2.02 Å Release Date: 2025-04-16 Classification: OXIDOREDUCTASE Ligands: GOL, HEM, TRP, TRS |
 |
Organism: Flavobacteriales bacterium
Method: X-RAY DIFFRACTION Resolution:2.51 Å Release Date: 2025-04-16 Classification: OXIDOREDUCTASE Ligands: HEM, IOP, GOL, TRS |

