Search Count: 723
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-09-17 Classification: DNA Ligands: K |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-09-17 Classification: DNA Ligands: K |
 |
 |
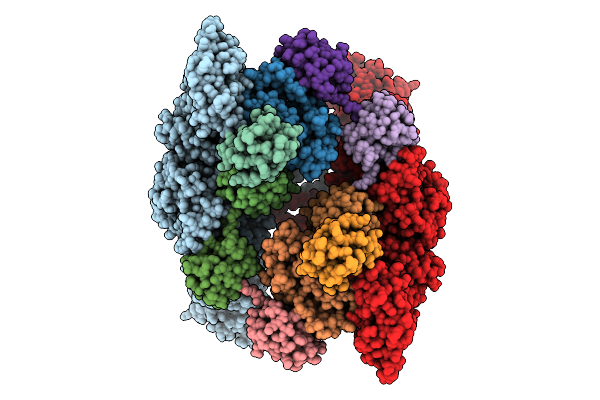
Cryo-Em Structure Of Ufd2/Ubc4-Ub In Complex With K29-Linked Diub (Monomeric Conformation)
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c), Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-07-30 Classification: LIGASE |
 |
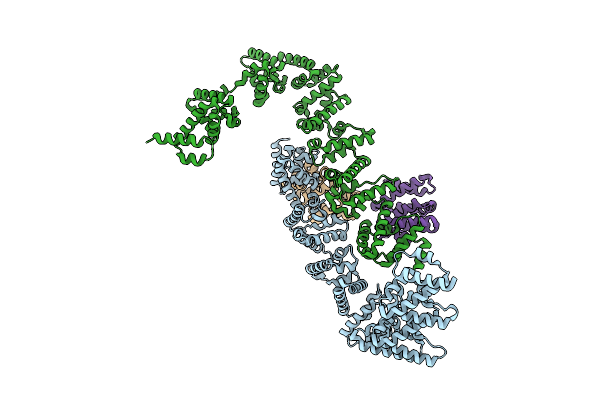
Cryo-Em Structure Of Ufd2/Ubc4-Ub In Complex With K29-Linked Triub (Dimeric Conformation)
Organism: Homo sapiens, Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: ELECTRON MICROSCOPY Release Date: 2025-07-30 Classification: LIGASE |
 |
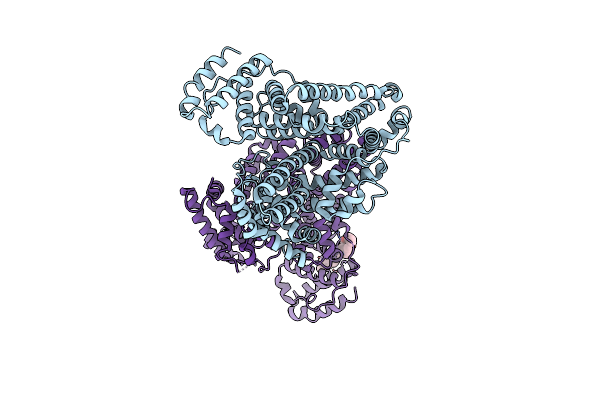
Crystal Structure Of Phytophthora Infestans Effector Avrcap1B In Complex With The Enth Domain Of Nicotiana Benthamiana Nbtol9A Protein
Organism: Phytophthora infestans, Nicotiana benthamiana
Method: X-RAY DIFFRACTION Release Date: 2025-07-16 Classification: PROTEIN BINDING |
 |
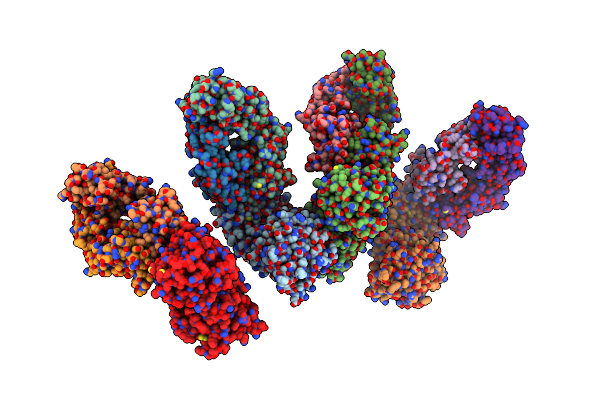
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-06-11 Classification: TRANSPORT PROTEIN Ligands: A1EGK |
 |
Crystal Structure Of N-Terminal Domain Of N-Methyl-D-Aspartate Receptor Subunit Nr1 In Complex With Patient-Derived Antibody
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-05-14 Classification: MEMBRANE PROTEIN |
 |
Organism: Thermoascus aurantiacus
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2025-04-30 Classification: HYDROLASE Ligands: MG, NOJ |
 |
Organism: Schizosaccharomyces pombe 972h-, Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-04-30 Classification: GENE REGULATION Ligands: SAM, ZN |
 |
Organism: Schizosaccharomyces pombe 972h-
Method: X-RAY DIFFRACTION Release Date: 2025-04-30 Classification: GENE REGULATION Ligands: SAM, ZN |
 |
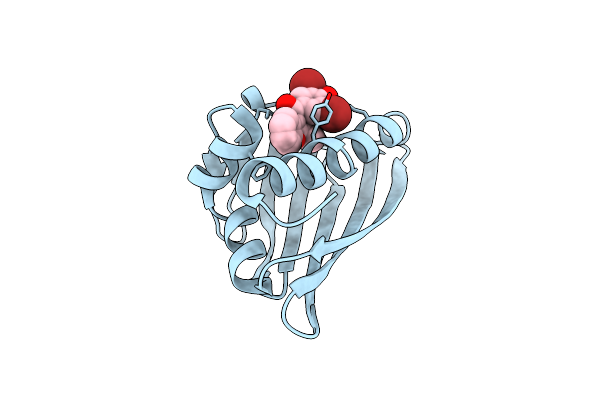
Benzbromarone Interferes With The Interaction Between Hsp90 And Aha1 By Interacting With Both Of Them
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-04-23 Classification: STRUCTURAL PROTEIN |
 |
Benzbromarone Interferes With The Interaction Between Hsp90 And Aha1 By Interacting With Both Of Them
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-04-23 Classification: STRUCTURAL PROTEIN Ligands: R75 |
 |
Glycoside Hydrolase Family 1 Beta-Glucosidase (E318G Mutant) From Streptomyces Griseus (Sophorose Complex)
Organism: Streptomyces griseus subsp. griseus nbrc 13350
Method: X-RAY DIFFRACTION Release Date: 2025-04-16 Classification: HYDROLASE Ligands: PGE, CL, BGC |
 |
Cryo-Em Structure Of The Sars-Cov-2 S 6P Trimer In Complex With The Human Neutralizing Antibody Fab Fragment Cav-C65 (Local Refinement)
Organism: Homo sapiens, Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2025-03-12 Classification: VIRAL PROTEIN |
 |
Cryo-Em Structure Of The Chemokine-Like Receptor 1 In Complex With Chemerin And Gi1
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-03-12 Classification: MEMBRANE PROTEIN |
 |
Cryo-Em Structure Of The G-Protein Coupled Receptor 1 (Gpr1) In Complex With Chemerin And Gi1
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-03-12 Classification: MEMBRANE PROTEIN |
 |
Organism: Mus musculus, Homo sapiens, Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2025-03-12 Classification: MEMBRANE PROTEIN/IMMUNE SYSTEM |
 |
Crystal Structure Of A Thioredoxin-Like Ferredoxin Of Rhodobacter Capsulatus
Organism: Rhodobacter capsulatus
Method: X-RAY DIFFRACTION Resolution:2.12 Å Release Date: 2025-02-19 Classification: ELECTRON TRANSPORT Ligands: FES |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2025-02-12 Classification: SUGAR BINDING PROTEIN Ligands: SER, GOL, PEG |

