Search Count: 189
 |
Organism: Homo sapiens, Dictyostelium discoideum
Method: X-RAY DIFFRACTION Release Date: 2025-09-24 Classification: MOTOR PROTEIN |
 |
Organism: Rhodococcus rhodochrous
Method: X-RAY DIFFRACTION Release Date: 2025-05-21 Classification: TRANSFERASE |
 |
Crystal Structure Of 4-Hydroxybenzoate-1-Hydroxylase From Gelatoporia Subvermispora (Gsmnx1)
Organism: Gelatoporia subvermispora
Method: X-RAY DIFFRACTION Resolution:1.82 Å Release Date: 2024-11-13 Classification: OXIDOREDUCTASE Ligands: FAD, CL |
 |
Crystal Structure Of Hydroquinone-2-Hydroxylase From Trametes Versicolor (Tvmnx3)
Organism: Trametes versicolor
Method: X-RAY DIFFRACTION Resolution:2.41 Å Release Date: 2024-11-13 Classification: OXIDOREDUCTASE Ligands: FAD |
 |
Crystal Structure Of Hydroxyquinol-1,2-Dioxygenase From Gelatoporia Subvermispora (Gshdx1)
Organism: Gelatoporia subvermispora
Method: X-RAY DIFFRACTION Resolution:1.78 Å Release Date: 2024-11-13 Classification: OXIDOREDUCTASE Ligands: SO4, PTY, FE |
 |
Crystal Structure Of Hydroxyquinol-1,2-Dioxygenase From Trametes Versicolor (Tvhdx1)
Organism: Trametes versicolor
Method: X-RAY DIFFRACTION Resolution:1.46 Å Release Date: 2024-11-13 Classification: OXIDOREDUCTASE Ligands: PTY, FE, TRS |
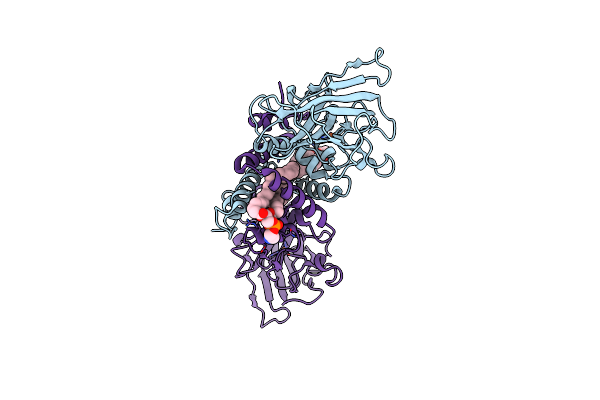 |
Crystal Structure Of Hydroxyquinol-1,2-Dioxygenase From Rhodococcus Jostii Rha1 (Rjtsdc)
Organism: Rhodococcus jostii rha1
Method: X-RAY DIFFRACTION Resolution:1.57 Å Release Date: 2024-11-13 Classification: OXIDOREDUCTASE Ligands: PTY, FE |
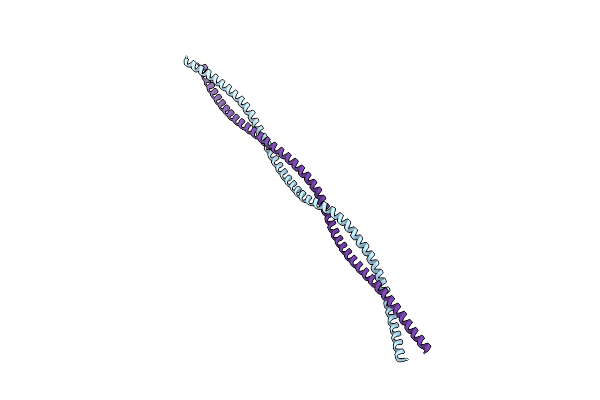 |
Organism: Schizosaccharomyces pombe
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2024-11-06 Classification: STRUCTURAL PROTEIN |
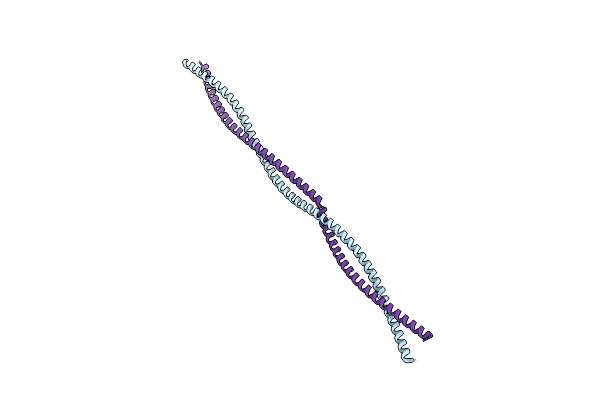 |
Organism: Schizosaccharomyces pombe
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2024-07-24 Classification: STRUCTURAL PROTEIN |
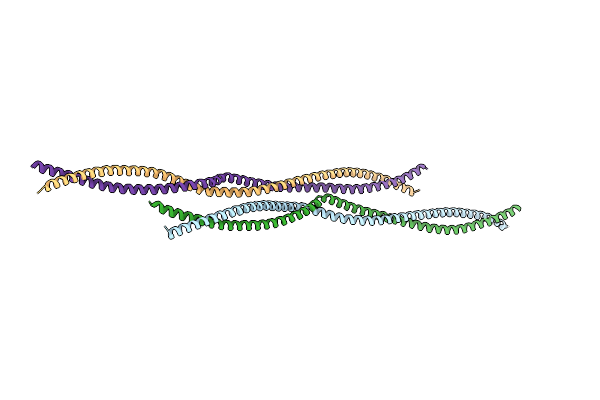 |
Organism: Schizosaccharomyces pombe
Method: X-RAY DIFFRACTION Resolution:2.43 Å Release Date: 2024-07-24 Classification: STRUCTURAL PROTEIN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-04-24 Classification: MEMBRANE PROTEIN |
 |
Crystal Structure Of Cutinase Adcut From Acidovorax Delafieldii (Pbs Depolymerase)
Organism: Acidovorax delafieldii
Method: X-RAY DIFFRACTION Resolution:1.49 Å Release Date: 2024-01-24 Classification: HYDROLASE |
 |
Organism: Rhizobacter gummiphilus
Method: X-RAY DIFFRACTION Resolution:1.08 Å Release Date: 2023-03-08 Classification: HYDROLASE Ligands: ACT |
 |
Organism: Pseudomonas saudimassiliensis
Method: X-RAY DIFFRACTION Resolution:1.56 Å Release Date: 2023-03-08 Classification: HYDROLASE Ligands: ACT |
 |
Organism: Halopseudomonas bauzanensis
Method: X-RAY DIFFRACTION Resolution:1.24 Å Release Date: 2023-03-08 Classification: HYDROLASE Ligands: SO4 |
 |
Crystal Structure Of The Malonyl-Acp Decarboxylase Madb From Pseudomonas Putida
Organism: Pseudomonas putida kt2440
Method: X-RAY DIFFRACTION Resolution:1.04 Å Release Date: 2023-03-01 Classification: TRANSFERASE |
 |
Organism: Novosphingobium aromaticivorans dsm 12444
Method: X-RAY DIFFRACTION Resolution:1.39 Å Release Date: 2023-02-01 Classification: LYASE Ligands: STL, SO4 |
 |
Organism: Novosphingobium aromaticivorans dsm 12444
Method: X-RAY DIFFRACTION Resolution:1.66 Å Release Date: 2023-02-01 Classification: LYASE Ligands: LJU |
 |
Organism: Sphingomonas paucimobilis
Method: X-RAY DIFFRACTION Resolution:1.68 Å Release Date: 2023-02-01 Classification: LYASE Ligands: GOL, SO4, LJU, LJL |
 |
Organism: Sphingobium sp. syk-6
Method: X-RAY DIFFRACTION Resolution:1.83 Å Release Date: 2023-02-01 Classification: LYASE Ligands: SO4, LJC, LHX |

