Search Count: 20
 |
Organism: Methylorubrum extorquens cm4
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2023-09-27 Classification: LYASE Ligands: MLT |
 |
Organism: Chloroflexus aurantiacus
Method: ELECTRON MICROSCOPY Release Date: 2023-08-30 Classification: OXIDOREDUCTASE |
 |
Crystal Structure Of Apo-Form Of Malonyl-Coa Reductase C-Domain From Chloroflexus Aurantiacus
Organism: Chloroflexus aurantiacus
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2023-06-14 Classification: OXIDOREDUCTASE Ligands: GOL, TLA |
 |
Crystal Structure Of Nadp-Binding Form Of Malonyl-Coa Reductase C-Domain From Chloroflexus Aurantiacus
Organism: Chloroflexus aurantiacus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2023-06-14 Classification: OXIDOREDUCTASE Ligands: GOL, TLA, NAP |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2020-05-13 Classification: OXIDOREDUCTASE Ligands: NO3, GOL, ZN |
 |
Structure Of Alcohol Dehydrogenase Yjgb In Complex With Nadp From Escherichia Coli
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2020-05-13 Classification: OXIDOREDUCTASE Ligands: ASP, NAP, GOL, PEG, ZN |
 |
Crystal Structure Of L-Aspartate/Glutamate-Specific Racemase From Escherichia Coli
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2015-11-18 Classification: ISOMERASE |
 |
Crystal Structure Of L-Aspartate/Glutamate Specific Racemase In Complex With L-Glutamate
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2015-11-18 Classification: ISOMERASE Ligands: GOL, GLU, NO3 |
 |
Crystal Structure Of Thiolase Mutation (V77Q,N153Y,A286K) From Clostridium Acetobutylicum
Organism: Clostridium acetobutylicum (strain atcc 824 / dsm 792 / jcm 1419 / lmg 5710 / vkm b-1787)
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2015-10-07 Classification: TRANSFERASE Ligands: GOL, PEG |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2015-10-07 Classification: TRANSFERASE |
 |
Crystal Structure Of Oxidized Form Of Thiolase From Clostridium Acetobutylicum
Organism: Clostridium acetobutylicum (strain atcc 824 / dsm 792 / jcm 1419 / lmg 5710 / vkm b-1787)
Method: X-RAY DIFFRACTION Resolution:1.77 Å Release Date: 2015-10-07 Classification: TRANSFERASE Ligands: ACT, PEG, GOL |
 |
Crystal Structure Of Reduced Form Of Thiolase From Clostridium Acetobutylicum
Organism: Clostridium acetobutylicum (strain ea 2018)
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2015-10-07 Classification: TRANSFERASE Ligands: GOL |
 |
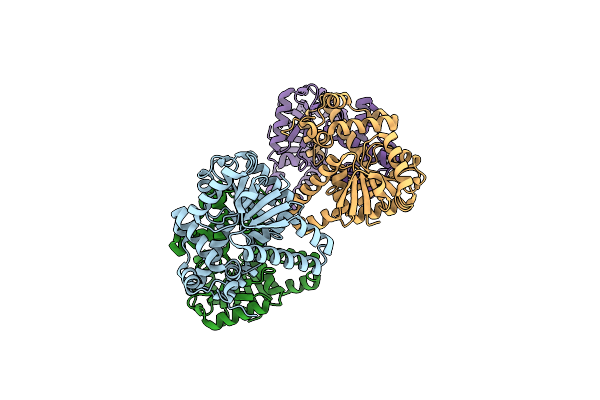
Crystal Structure Of Thiolase From Clostridium Acetobutylicum In Complex With Coa
Organism: Clostridium acetobutylicum (strain ea 2018)
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2015-10-07 Classification: TRANSFERASE Ligands: COA, GOL |
 |
Crystal Structure Of (S)-3-Hydroxybutylryl-Coa Dehydrogenase Form The N-Butanol Sysnthesizing Bacterium, Clostridium Butyricum.
Organism: Clostridium butyricum e4 str. bont e bl5262
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2015-07-29 Classification: OXIDOREDUCTASE |
 |
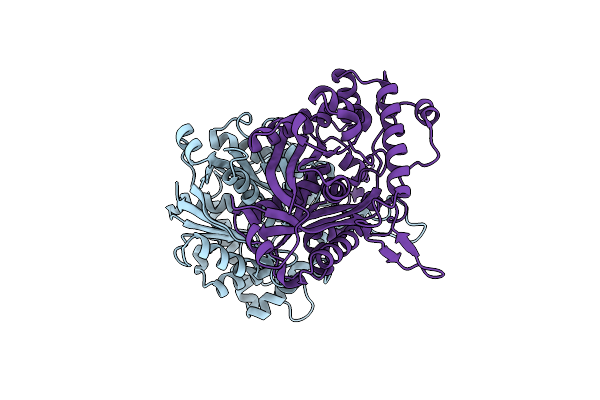
Organism: Nonlabens dokdonensis dsw-6
Method: X-RAY DIFFRACTION Resolution:1.97 Å Release Date: 2015-07-15 Classification: OXIDOREDUCTASE, FLAVOPROTEIN Ligands: FAD, GOL |
 |
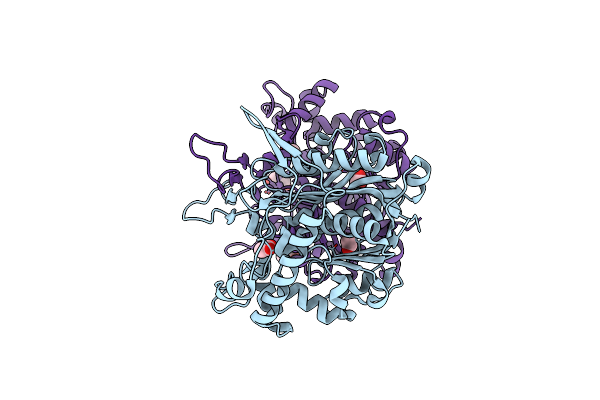
Organism: Ralstonia eutropha
Method: X-RAY DIFFRACTION Resolution:2.29 Å Release Date: 2014-11-19 Classification: TRANSFERASE |
 |
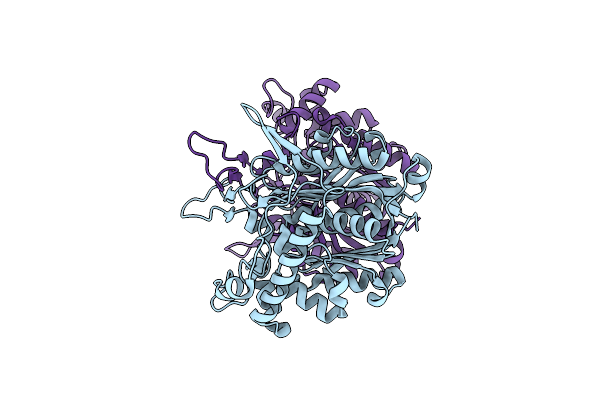
Crystal Structure Of Oxidized Form Of Thiolase From Clostridium Acetobutylicum
Organism: Clostridium acetobutylicum
Method: X-RAY DIFFRACTION Resolution:1.77 Å Release Date: 2014-10-08 Classification: TRANSFERASE Ligands: ACT, GOL |
 |
Crystal Structure Of Reduced Form Of Thiolase From Clostridium Acetobutylicum
Organism: Clostridium acetobutylicum
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2014-10-08 Classification: TRANSFERASE |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.74 Å Release Date: 2014-03-12 Classification: GENE REGULATION |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.92 Å Release Date: 2014-03-12 Classification: GENE REGULATION |

