Search Count: 19
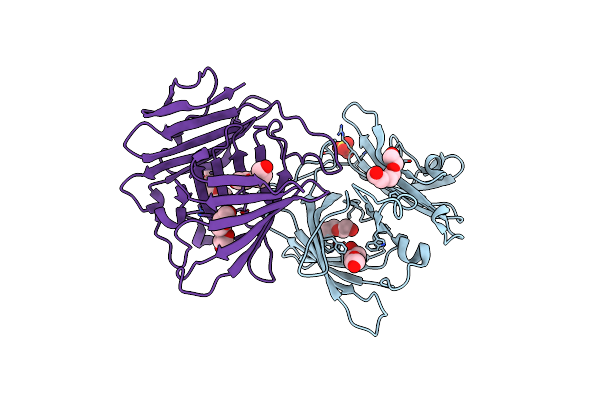 |
Crystal Structure Of Uncharacterized Protein Ylr301W From Saccharomycces Cerevisiae
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2012-03-21 Classification: UNKNOWN FUNCTION Ligands: PGE, SO4 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2008-10-14 Classification: CHAPERONE Ligands: PO4 |
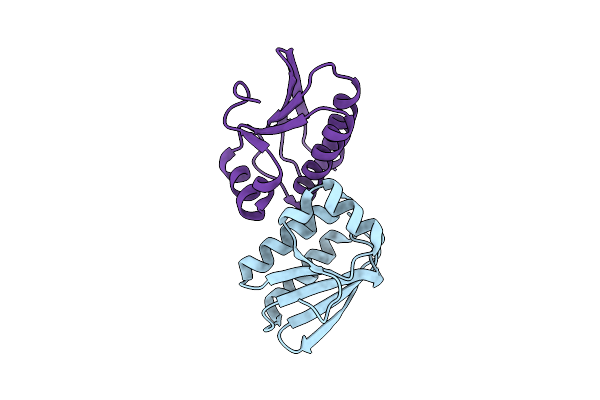 |
Crystal Structure Of The Tm1442 Protein From Thermotoga Maritima, A Homolog Of The Bacillus Subtilis General Stress Response Anti-Anti-Sigma Factor Rsbv
Organism: Thermotoga maritima
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2004-09-28 Classification: GENE REGULATION |
 |
Crystal Structure Of Agmatinase Reveals Structural Conservation And Inhibition Mechanism Of The Ureohydrolase Superfamily
Organism: Deinococcus radiodurans
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2004-09-07 Classification: HYDROLASE Ligands: MN, 16D |
 |
Crystal Structure Of Agmatinase Reveals Structural Conservation And Inhibition Mechanism Of The Ureohydrolase Superfamily
Organism: Deinococcus radiodurans
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2004-09-07 Classification: HYDROLASE |
 |
Crystal Structure Of Agmatinase Reveals Structural Conservation And Inhibition Mechanism Of The Ureohydrolase Superfamily
Organism: Deinococcus radiodurans
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2004-09-07 Classification: HYDROLASE Ligands: MN |
 |
Organism: Thermotoga maritima
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2004-09-07 Classification: TRANSFERASE Ligands: THG |
 |
Organism: Thermotoga maritima
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2004-09-07 Classification: TRANSFERASE Ligands: FFO |
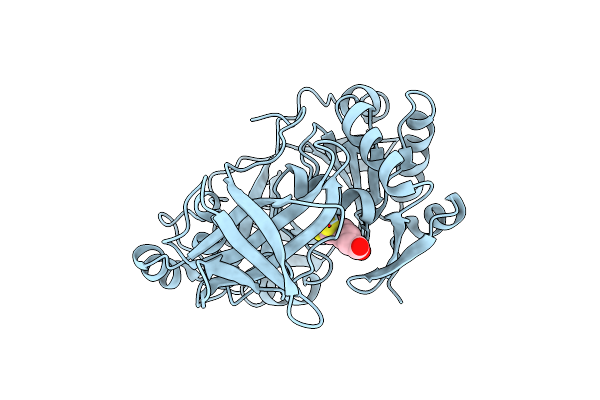 |
Organism: Thermotoga maritima
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2004-09-07 Classification: TRANSFERASE Ligands: RED |
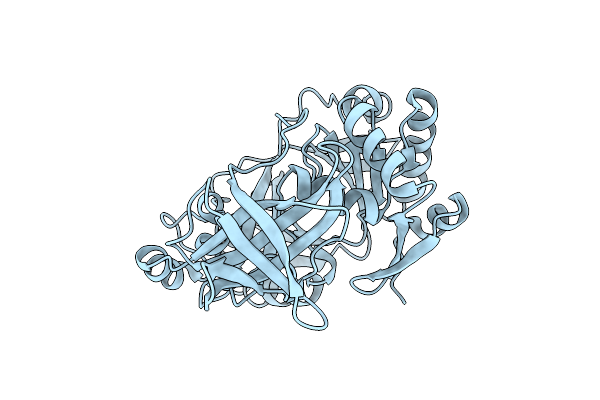 |
Organism: Thermotoga maritima
Method: X-RAY DIFFRACTION Resolution:1.84 Å Release Date: 2004-09-07 Classification: TRANSFERASE |
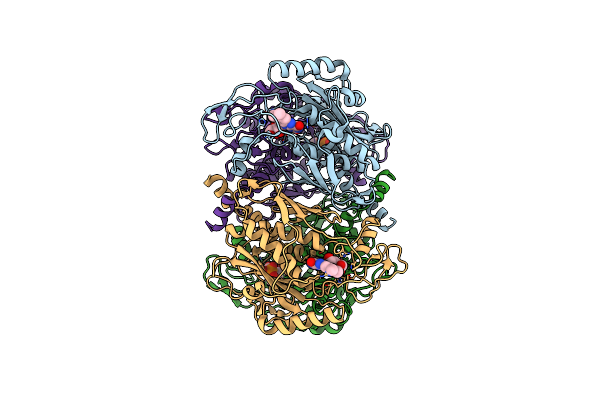 |
Organism: Helicobacter pylori
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2004-06-01 Classification: LYASE Ligands: FMN |
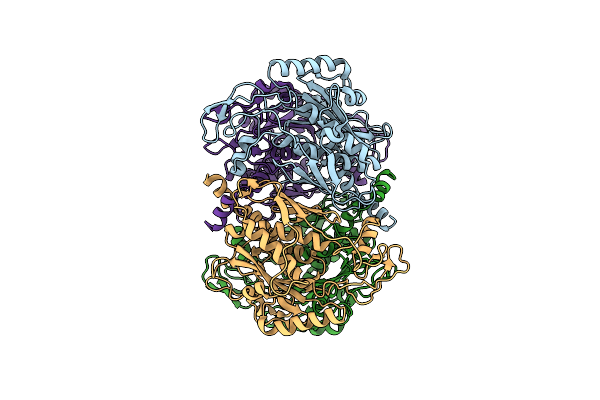 |
Organism: Helicobacter pylori
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2004-06-01 Classification: LYASE |
 |
Crystal Structure Of Beta-D-Xylosidase From Thermoanaerobacterium Saccharolyticum, A Family 39 Glycoside Hydrolase
Organism: Thermoanaerobacterium saccharolyticum
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2003-12-23 Classification: HYDROLASE Ligands: XYP |
 |
Crystal Structure Of Beta-D-Xylosidase From Thermoanaerobacterium Saccharolyticum, A Family 39 Glycoside Hydrolase
Organism: Thermoanaerobacterium saccharolyticum
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2003-12-23 Classification: HYDROLASE Ligands: DFX |
 |
Crystal Structure Of Trna(M1G37)Methyltransferase: Insight Into Trna Recognition
Organism: Haemophilus influenzae
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2003-06-17 Classification: TRANSFERASE |
 |
Crystal Structure Of Trna(M1G37)Methyltransferase: Insight Into Trna Recognition
Organism: Haemophilus influenzae
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2003-06-17 Classification: TRANSFERASE Ligands: SAM |
 |
Crystal Structure Of Trna(M1G37)Methyltransferase: Insight Into Trna Recognition
Organism: Haemophilus influenzae
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2003-06-17 Classification: TRANSFERASE Ligands: SAH |
 |
Crystal Structure Of Trna(M1G37)Methyltransferase: Insight Into Trna Recognition
Organism: Haemophilus influenzae
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2003-06-17 Classification: TRANSFERASE Ligands: PO4, SAH |
 |
Crystal Structure Of Class I Acetohydroxy Acid Isomeroreductase From Pseudomonas Aeruginosa
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2003-05-06 Classification: OXIDOREDUCTASE |

