Search Count: 52
 |
Set Domain Of Histone-Lysine N-Methyltransferase Nsd2 In Complex With Selective Inhibitor
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 2025-04-30 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: ZN, A1A0M |
 |
Molecular Docking Of Spf30 Tudor Domain With Synthetic Inhibitor 4-(Pyridin-2-Yl)Thiazol-2-Amine
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 2023-08-23 Classification: RNA BINDING PROTEIN Ligands: ZTI |
 |
Organism: Methylococcus capsulatus
Method: ELECTRON MICROSCOPY Release Date: 2023-01-25 Classification: OXIDOREDUCTASE Ligands: FE |
 |
Organism: Methylococcus capsulatus
Method: ELECTRON MICROSCOPY Release Date: 2023-01-25 Classification: OXIDOREDUCTASE Ligands: FE |
 |
Crystal Structure Of Penicillium Verruculosum Copalyl Diphosphate Synthase (Pvcps) Alpha Prenyltransferase Domain Variant, F760A
Organism: Talaromyces verruculosus
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2021-10-13 Classification: TRANSFERASE |
 |
Crystal Structure Of Penicillium Verruculosum Copalyl Diphosphate Synthase (Pvcps) Alpha Prenyltransferase Domain Variant, H786A
Organism: Talaromyces verruculosus
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2021-10-13 Classification: TRANSFERASE |
 |
Crystal Structure Of Penicillium Verruculosum Copalyl Diphosphate Synthase (Pvcps) Alpha Prenyltransferase Domain Variant, S723Y
Organism: Talaromyces verruculosus
Method: X-RAY DIFFRACTION Resolution:3.15 Å Release Date: 2021-10-13 Classification: TRANSFERASE |
 |
Crystal Structure Of Penicillium Verruculosum Copalyl Diphosphate Synthase (Pvcps) Alpha Prenyltransferase Domain Variant, S723F
Organism: Talaromyces verruculosus
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2021-10-13 Classification: TRANSFERASE |
 |
Crystal Structure Of Penicillium Verruculosum Copalyl Diphosphate Synthase (Pvcps) Alpha Prenyltransferase Domain Variant, S723T, Bound With Non-Productive Isopentenyl Diphosphate
Organism: Talaromyces verruculosus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2021-10-13 Classification: TRANSFERASE Ligands: IPE |
 |
Discovery Of A Dual Inhibitor Of Nqo1 And Gstp1 For Treating Malignant Glioblastoma
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2020-11-25 Classification: OXIDOREDUCTASE Ligands: FAD, EHL, SO4 |
 |
Discovery Of A Dual Inhibitor Of Nqo1 And Gstp1 For Treating Malignant Glioblastoma
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.58 Å Release Date: 2020-11-25 Classification: TRANSFERASE Ligands: GSH, MES |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.25 Å Release Date: 2020-08-12 Classification: BIOSYNTHETIC PROTEIN Ligands: EDO |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.15 Å Release Date: 2020-08-12 Classification: BIOSYNTHETIC PROTEIN Ligands: EDO |
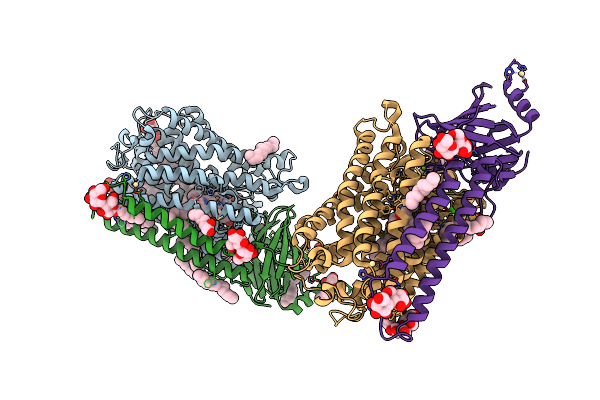 |
Organism: Rhodobacter sphaeroides 2.4.1
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2019-11-27 Classification: OXIDOREDUCTASE Ligands: OH, HEA, DMU, TRD, HTH, CU, MG, CA, CD, TRS |
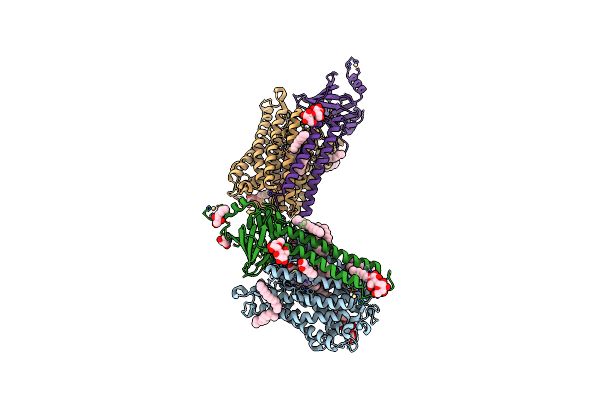 |
Organism: Rhodobacter sphaeroides 2.4.1
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2019-11-27 Classification: OXIDOREDUCTASE Ligands: TRD, DMU, HEA, CU, MG, CA, HTH, TRS, CD |
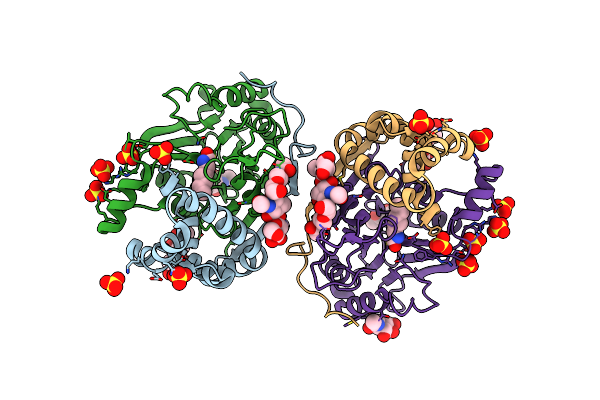 |
Crystal Structure Of Human Acid Ceramidase In Covalent Complex With Carmofur
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.74 Å Release Date: 2019-01-23 Classification: HYDROLASE Ligands: SO4, NAG, JRY |
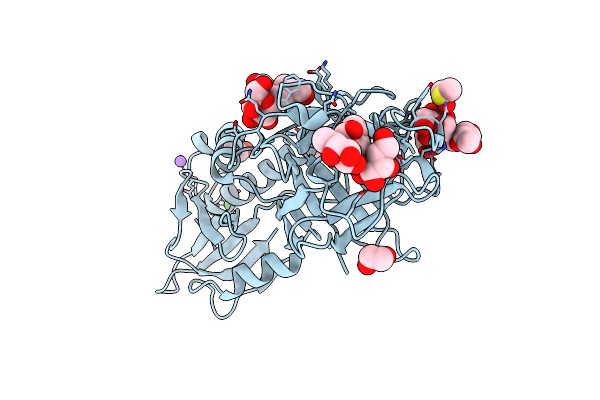 |
Organism: Marasmius oreades, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2016-03-02 Classification: HYDROLASE Ligands: EDO, CL, CA, DMS, NA |
 |
Organism: Marasmius oreades, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2016-03-02 Classification: HYDROLASE Ligands: FUC, EDO, DMS, CL, CA |
 |
Organism: Marasmius oreades, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2016-03-02 Classification: HYDROLASE Ligands: EDO, DMS, CL, CA, NA |
 |
Organism: Lyophyllum decastes
Method: X-RAY DIFFRACTION Resolution:1.00 Å Release Date: 2014-12-10 Classification: SUGAR BINDING PROTEIN |

