Search Count: 26
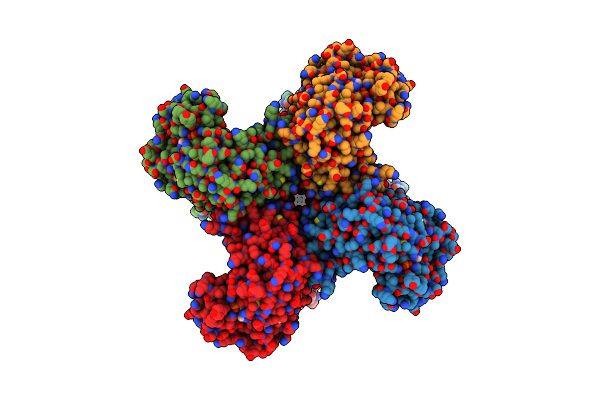 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2022-02-09 Classification: MEMBRANE PROTEIN Ligands: K, NAP |
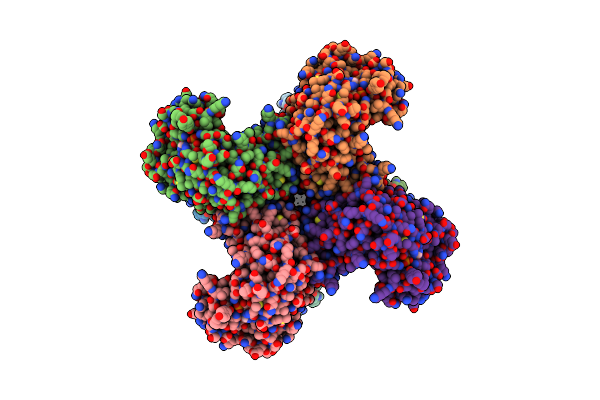 |
Composite Map Of Human Kv1.3 Channel In Dalazatide-Bound State With Beta Subunits
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2022-02-09 Classification: MEMBRANE PROTEIN Ligands: NAP, K |
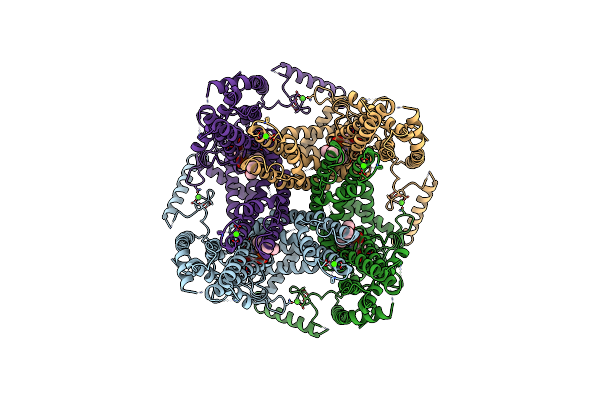 |
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2021-04-21 Classification: MEMBRANE PROTEIN Ligands: CA, PWE |
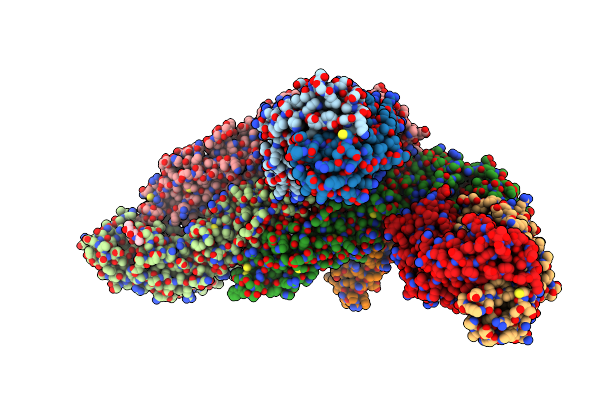 |
Organism: Homo sapiens, Zika virus
Method: ELECTRON MICROSCOPY Release Date: 2020-07-08 Classification: VIRUS |
 |
Organism: Mycobacterium smegmatis str. mc2 155, Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2018-09-26 Classification: RIBOSOME |
 |
Organism: Mycobacterium smegmatis str. mc2 155, Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2018-09-26 Classification: RIBOSOME |
 |
Organism: Mycobacterium smegmatis str. mc2 155
Method: ELECTRON MICROSCOPY Release Date: 2018-09-26 Classification: RIBOSOME |
 |
Organism: Mycobacterium smegmatis (strain atcc 700084 / mc(2)155), Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2018-09-26 Classification: RIBOSOME |
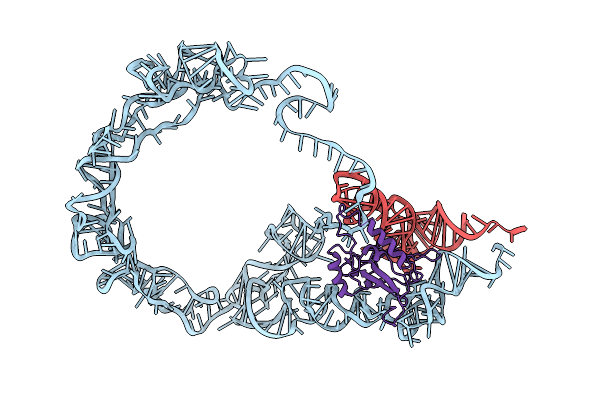 |
Organism: Mycobacterium smegmatis (strain atcc 700084 / mc(2)155), Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2018-09-26 Classification: RNA BINDING PROTEIN/RNA |
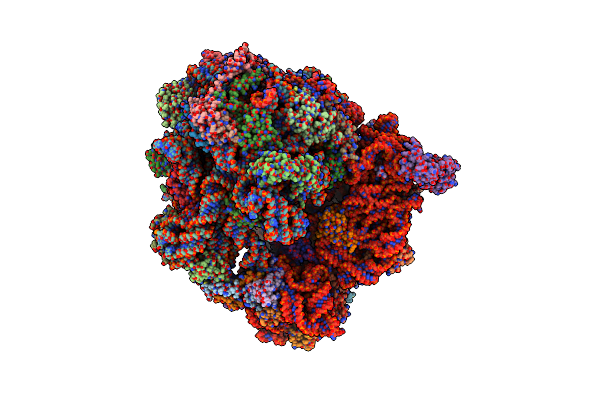 |
Organism: Spinacia oleracea
Method: ELECTRON MICROSCOPY Release Date: 2017-06-14 Classification: RIBOSOME |
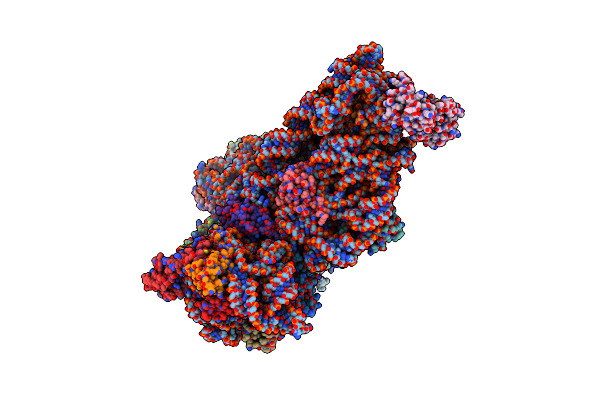 |
Organism: Spinacia oleracea
Method: ELECTRON MICROSCOPY Release Date: 2017-06-07 Classification: RIBOSOME |
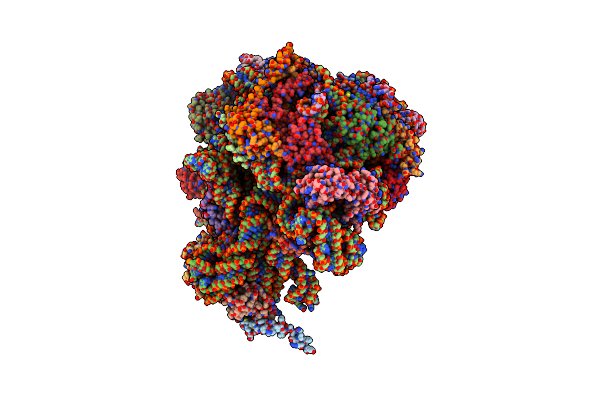 |
Organism: Spinacia oleracea
Method: ELECTRON MICROSCOPY Release Date: 2017-06-07 Classification: RIBOSOME |
 |
Methicillin Resistant, Linezolid Resistant Staphylococcus Aureus 70S Ribosome (Delta S145 Ul3)
Organism: Staphylococcus aureus
Method: ELECTRON MICROSCOPY Release Date: 2017-05-31 Classification: RIBOSOME Ligands: MG |
 |
Organism: Staphylococcus aureus subsp. aureus nctc 8325, Staphylococcus aureus (strain nctc 8325)
Method: ELECTRON MICROSCOPY Release Date: 2017-05-24 Classification: ribosome/antibiotic Ligands: PAR, MG |
 |
Organism: Spinacia oleracea
Method: ELECTRON MICROSCOPY Release Date: 2017-02-01 Classification: RIBOSOME |
 |
Crystal Structure Of The Braf (R509H) Kinase Domain Monomer Bound To Vemurafenib
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.99 Å Release Date: 2016-08-10 Classification: Transferase/Transferase Inhibitor Ligands: 032 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.49 Å Release Date: 2016-08-10 Classification: Transferase/Transferase Inhibitor Ligands: B1E |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2016-08-10 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: 6DC |
 |
Organism: Thermus thermophilus (strain hb8 / atcc 27634 / dsm 579), Salmonella enterica subsp. enterica serovar enteritidis str. sa20094389
Method: ELECTRON MICROSCOPY Release Date: 2016-05-18 Classification: RIBOSOME Ligands: GCP |
 |
Organism: Thermus thermophilus (strain hb8 / atcc 27634 / dsm 579), Salmonella enterica subsp. enterica serovar enteritidis str. sa20094389
Method: ELECTRON MICROSCOPY Release Date: 2016-05-18 Classification: RIBOSOME Ligands: GCP |

