Search Count: 7
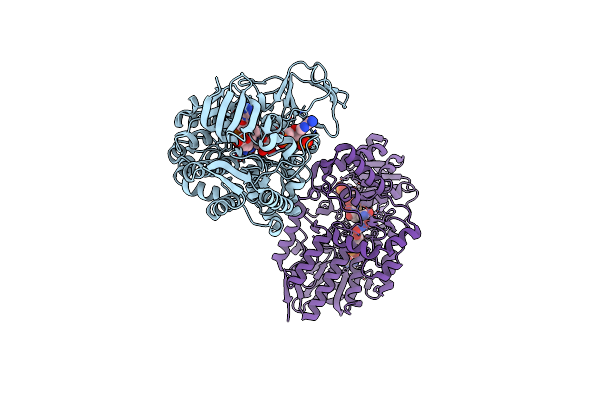 |
Crystal Structure Of Cyclohexanone Monooxygenase From Rhodococcus Sp. Phi1 Bound To Nadp+
Organism: Rhodococcus sp. phi1
Method: X-RAY DIFFRACTION Resolution:2.37 Å Release Date: 2018-09-26 Classification: FLAVOPROTEIN Ligands: FAD, NAP |
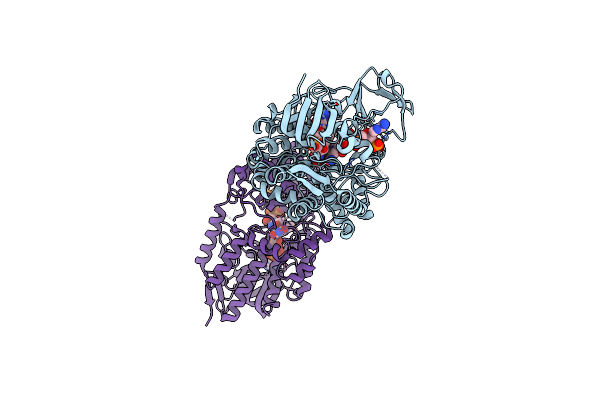 |
Crystal Structure Of Cyclohexanone Monooxygenase Mutant (F249A, F280A And F435A) From Rhodococcus Sp. Phi1 Bound To Nadp+
Organism: Rhodococcus sp. phi1
Method: X-RAY DIFFRACTION Release Date: 2018-09-26 Classification: FLAVOPROTEIN Ligands: FAD, NAP |
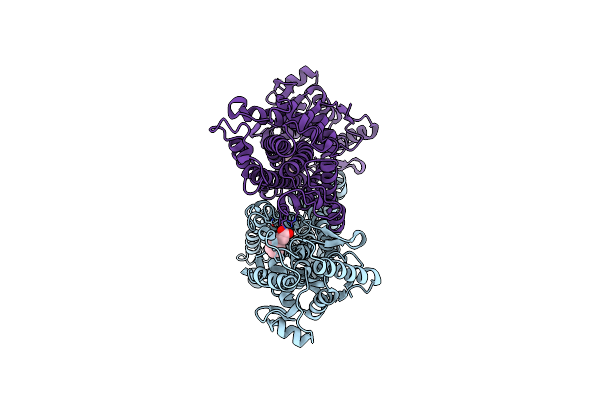 |
Crystal Structure Of Phenylalanine Ammonia-Lyase From Anabaena Variabilis (Y78F-C503S-C565S) Bound To Cinnamate
Organism: Anabaena
Method: X-RAY DIFFRACTION Resolution:2.41 Å Release Date: 2017-09-20 Classification: LYASE Ligands: HCI |
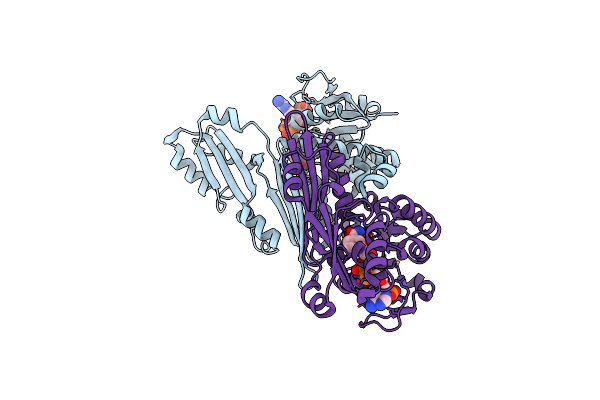 |
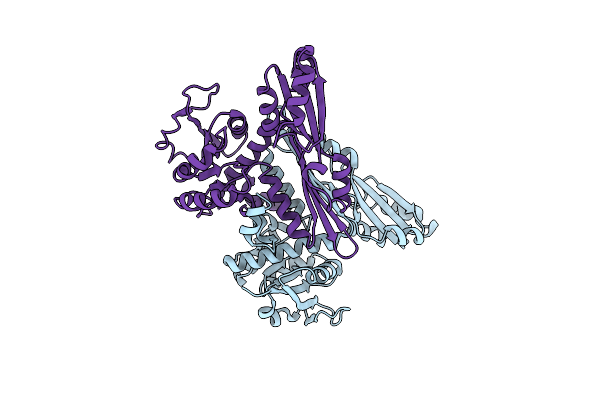
Crystal Structure Of The Engineered D-Amino Acid Dehydrogenase (Daadh) Bound To Nadp+
Organism: Corynebacterium glutamicum (strain atcc 13032 / dsm 20300 / jcm 1318 / lmg 3730 / ncimb 10025)
Method: X-RAY DIFFRACTION Resolution:1.84 Å Release Date: 2017-08-16 Classification: OXIDOREDUCTASE Ligands: NAP |
 |
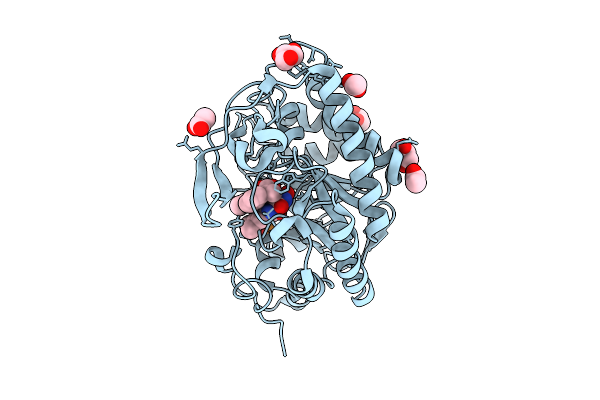
Organism: Corynebacterium glutamicum
Method: X-RAY DIFFRACTION Resolution:2.04 Å Release Date: 2017-08-16 Classification: OXIDOREDUCTASE |
 |
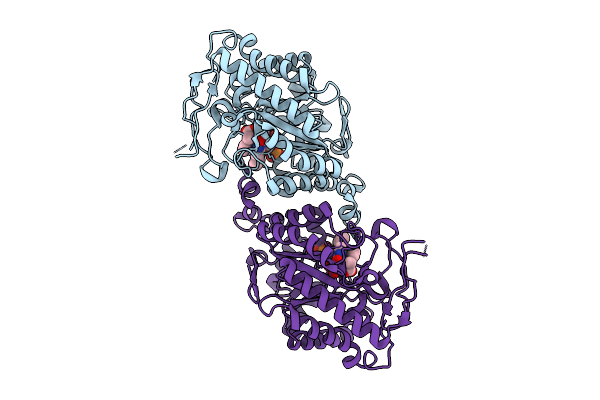
Organism: Agrobacterium tumefaciens
Method: X-RAY DIFFRACTION Resolution:1.58 Å Release Date: 2017-05-17 Classification: OXIDOREDUCTASE Ligands: FMN, 8OZ, GOL, ACT |
 |
Xenobiotic Reductase A (Xena) From Pseudomonas Putida In Complex With 2-Phenylacrylic Acid
Organism: Pseudomonas putida
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2017-05-17 Classification: OXIDOREDUCTASE Ligands: 8OZ, CL, GOL, FMN |

