Search Count: 20
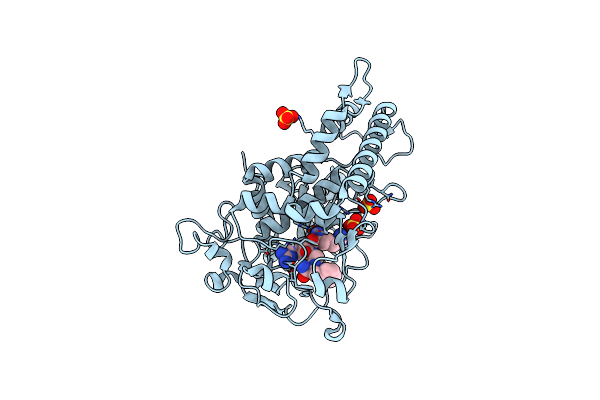 |
Crystal Structure Of The Protease Domain Of Botulinum Neurotoxin Serotype A With A Peptide Inhibitor Rrgf
Organism: Clostridium botulinum
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2012-02-08 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: ZN, SO4 |
 |
Crystal Structure Of The Protease Domain Of Botulinum Neurotoxin Serotype A With A Peptide Inhibitor Rygc
Organism: Clostridium botulinum
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2012-02-08 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: ZN, SO4, NA |
 |
Crystal Structure Of The Protease Domain Of Botulinum Neurotoxin Serotype A With A Peptide Inhibitor Rrfc
Organism: Clostridium botulinum
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2012-02-08 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: ZN, SO4, NA |
 |
Crystal Structure Of The Protease Domain Of Botulinum Neurotoxin Serotype A With A Peptide Inhibitor Crgc
Organism: Clostridium botulinum
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2012-02-08 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: ZN, SO4, NA, EDO |
 |
Crystal Structure Of The Catalytic Domain Of Botulinum Neurotoxin Serotype A With A Snap-25 Peptide
Organism: Clostridium botulinum, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2008-09-16 Classification: HYDROLASE Ligands: ZN, SO4 |
 |
Crystal Structure Of The Catalytic Domain Of Botulinum Neurotoxin Serotype A With A Substrate Analog Peptide
Organism: Clostridium botulinum
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2008-09-16 Classification: HYDROLASE Ligands: ZN, SO4 |
 |
Crystal Structure Of The Catalytic Domain Of Botulinum Neurotoxin Serotype A With An Acetate Ion Bound At The Active Site
Organism: Clostridium botulinum
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2008-04-22 Classification: HYDROLASE Ligands: ZN, SO4, ACT |
 |
Crystal Structure Of The Catalytic Domain Of Botulinum Neurotoxin Serotype A With Inhibitory Peptide Rrgc
Organism: Clostridium botulinum
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2008-04-22 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: ZN, SO4, NA |
 |
Crystal Structure Of The Catalytic Domain Of Botulinum Neurotoxin Serotype A With Inhibitory Peptide Rrgm
Organism: Clostridium botulinum
Method: X-RAY DIFFRACTION Resolution:1.58 Å Release Date: 2008-04-22 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: ZN, SO4 |
 |
Crystal Structure Of The Catalytic Domain Of Botulinum Neurotoxin Serotype A With Inhibitory Peptide Rrgl
Organism: Clostridium botulinum, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.52 Å Release Date: 2008-04-22 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: ZN, SO4 |
 |
Crystal Structure Of The Catalytic Domain Of Botulinum Neurotoxin Serotype A With Inhibitory Peptide Rrgi
Organism: Clostridium botulinum, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.47 Å Release Date: 2008-04-22 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: ZN, SO4 |
 |
N-Terminal Helix Reorients In Recombinant C-Fragment Of Clostridium Botulinum Type B
Organism: Clostridium botulinum
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2005-03-15 Classification: HYDROLASE |
 |
Organism: Salmonella typhimurium
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 1999-03-23 Classification: LYASE |
 |
Crystal Structures Of Mutant (Betak87T) Tryptophan Synthase Alpha2 Beta2 Complex With Ligands Bound To The Active Sites Of The Alpha And Beta Subunits Reveal Ligand-Induced Conformational Changes
Organism: Salmonella typhimurium
Method: X-RAY DIFFRACTION Resolution:2.04 Å Release Date: 1997-04-01 Classification: LYASE Ligands: IPL, NA, PLS |
 |
Crystal Structures Of Mutant (Betak87T) Tryptophan Synthase Alpha2 Beta2 Complex With Ligands Bound To The Active Sites Of The Alpha And Beta Subunits Reveal Ligand-Induced Conformational Changes
Organism: Salmonella typhimurium
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 1997-04-01 Classification: LYASE Ligands: G3P, NA, PLS |
 |
Crystal Structures Of Mutant (Betak87T) Tryptophan Synthase Alpha2 Beta2 Complex With Ligands Bound To The Active Sites Of The Alpha And Beta Subunits Reveal Ligand-Induced Conformational Changes
Organism: Salmonella typhimurium
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 1997-04-01 Classification: LYASE |
 |
Tryptophan Synthase (E.C.4.2.1.20) In The Presence Of Cesium, Room Temperature
Organism: Salmonella typhimurium
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 1996-03-08 Classification: CARBON-OXYGEN LYASE Ligands: CS, PLP |
 |
Tryptophan Synthase (E.C.4.2.1.20) In The Presence Of Potassium At Room Temperature
Organism: Salmonella typhimurium
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 1996-03-08 Classification: CARBON-OXYGEN LYASE Ligands: K, PLP |
 |
Tryptophan Synthase (E.C.4.2.1.20) With A Mutation Of Lys 87->Thr In The B Subunit And In The Presence Of Ligand L-Serine
Organism: Salmonella typhimurium
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 1996-03-08 Classification: LYASE/PEPTIDE |
 |
Structure Of Nadh Peroxidase From Streptococcus Faecalis 10C1 Refined At 2.16 Angstroms Resolution
Organism: Enterococcus faecalis
Method: X-RAY DIFFRACTION Resolution:2.16 Å Release Date: 1994-01-31 Classification: OXIDOREDUCTASE(H2O2(A)) Ligands: FAD |

