Search Count: 18
 |
Organism: Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-08-20 Classification: DNA |
 |
Organism: Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-08-20 Classification: DNA BINDING PROTEIN Ligands: ATP, ADP, ZN |
 |
Organism: Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-08-20 Classification: DNA BINDING PROTEIN |
 |
Organism: Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-08-20 Classification: DNA BINDING PROTEIN Ligands: ADP, ATP |
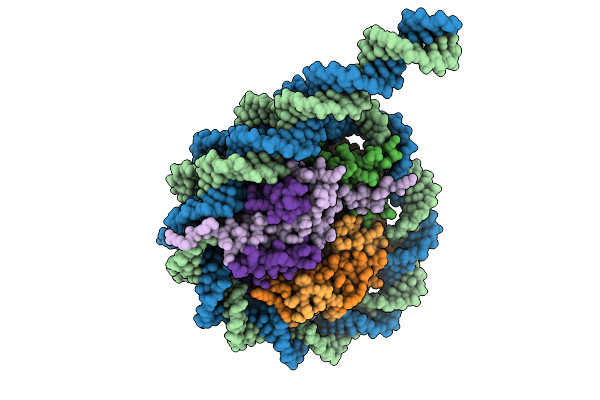 |
Organism: Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-08-20 Classification: DNA BINDING PROTEIN |
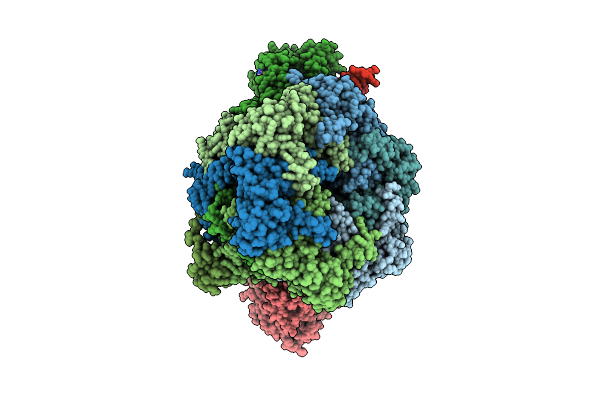 |
Organism: Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-08-20 Classification: DNA BINDING PROTEIN Ligands: ATP, ADP, ZN |
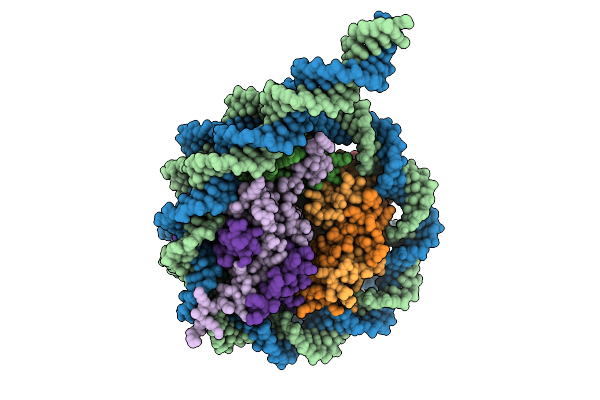 |
Organism: Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-08-20 Classification: DNA BINDING PROTEIN |
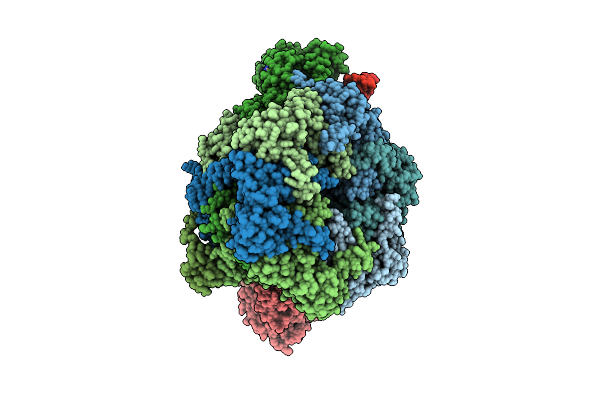 |
Organism: Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-08-13 Classification: DNA BINDING PROTEIN Ligands: ADP, ATP, ZN |
 |
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 2021-05-12 Classification: DNA BINDING PROTEIN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.57 Å Release Date: 2021-03-17 Classification: DNA BINDING PROTEIN/DNA Ligands: SO4 |
 |
Structure Of [T9,K7]Indolicidin, A Non Glycosylated Analogue Of Indolicidin
Organism: Bos taurus
Method: SOLUTION NMR Release Date: 2019-06-19 Classification: ANTIMICROBIAL PROTEIN |
 |
Structure Of [Beta Glc-T9,K7]Indolicidin, A Glycosylated Analogue Of Indolicidin
Organism: Bos taurus
Method: SOLUTION NMR Release Date: 2019-06-19 Classification: ANTIMICROBIAL PROTEIN Ligands: BGC |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:3.06 Å Release Date: 2009-09-01 Classification: HYDROLASE Ligands: MG, CL |
 |
Crystal Structure Of Yrbi Lacking The Last 8 Residues, In Complex With Kdo And Inorganic Phosphate
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.49 Å Release Date: 2009-09-01 Classification: HYDROLASE Ligands: MG, PO4, KDO |
 |
Organism: Escherichia coli o6
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2008-09-23 Classification: HYDROLASE Ligands: MG, CL |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2008-09-23 Classification: HYDROLASE Ligands: CL |
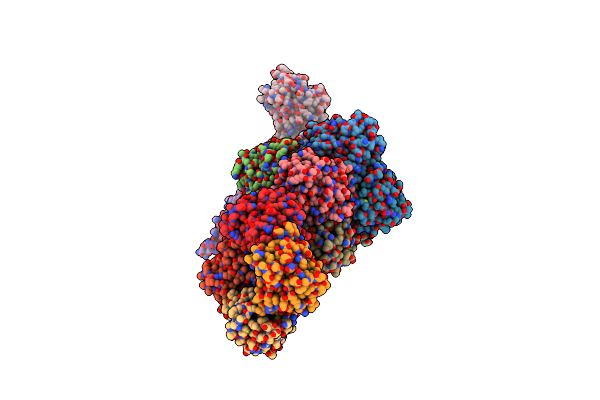 |
Crystal Structure Of Yrbi Phosphatase From Escherichia Coli In A Complex With Ca
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2008-09-23 Classification: HYDROLASE Ligands: CA, CL |
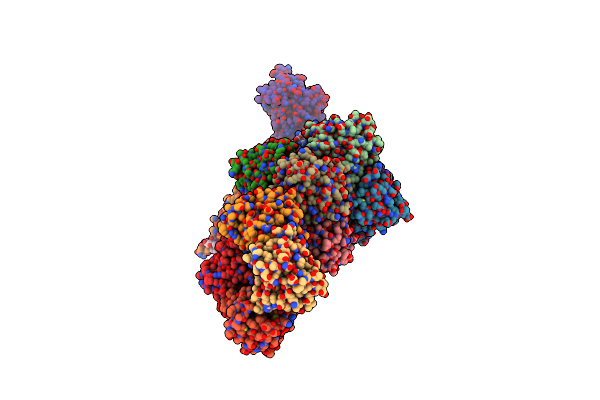 |
Crystal Structure Of Yrbi Phosphatase From Escherichia Coli In Complex With A Phosphate And A Calcium Ion
Organism: Escherichia coli o6
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2008-09-23 Classification: HYDROLASE Ligands: CA, PO4 |

