Search Count: 167
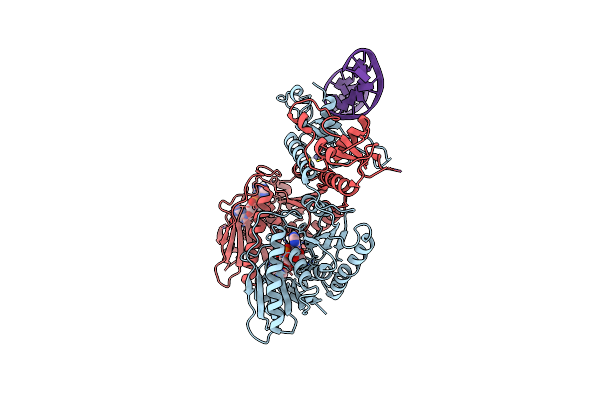 |
Cbass Pseudomonas Syringae Cap5 Tetramer With Dna Duplex And 3'2'-C-Gamp Cyclic Dinucleotide Ligand
Organism: Pseudomonas syringae, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.67 Å Release Date: 2025-06-18 Classification: IMMUNE SYSTEM Ligands: 4UR, ZN |
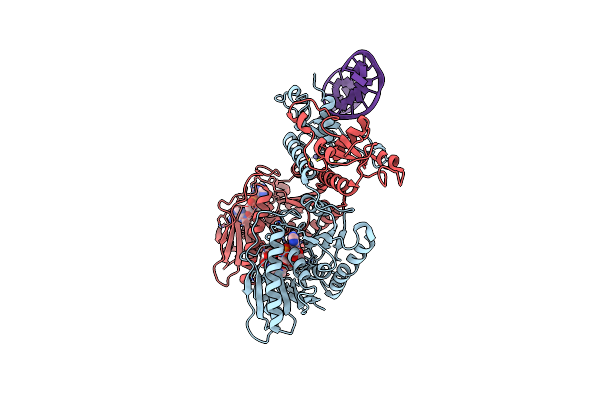 |
Cbass Pseudomonas Syringae Cap5 Tetramer With Dna Duplex And 3'2'-C-Diamp Cyclic Dinucleotide Ligand
Organism: Pseudomonas syringae, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.94 Å Release Date: 2025-06-18 Classification: IMMUNE SYSTEM Ligands: ZN, Y4F, GOL |
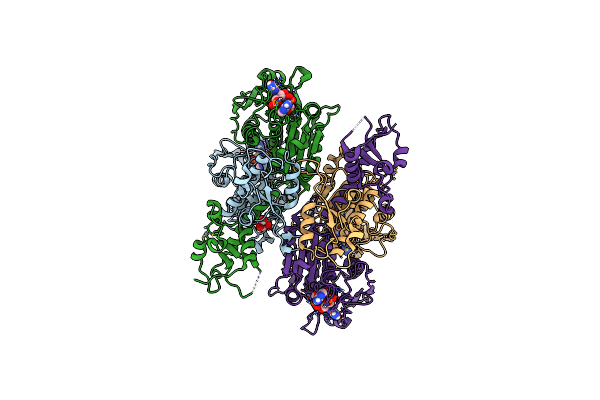 |
Cbass Pseudomonas Syringae Cap5 Tetramer With 3'2'-C-Gamp Cyclic Dinucleotide Ligand (His56Ala Mutant Without Mg2+ Ions)
Organism: Pseudomonas syringae
Method: X-RAY DIFFRACTION Resolution:1.64 Å Release Date: 2025-06-18 Classification: IMMUNE SYSTEM Ligands: ZN, 4UR, GOL |
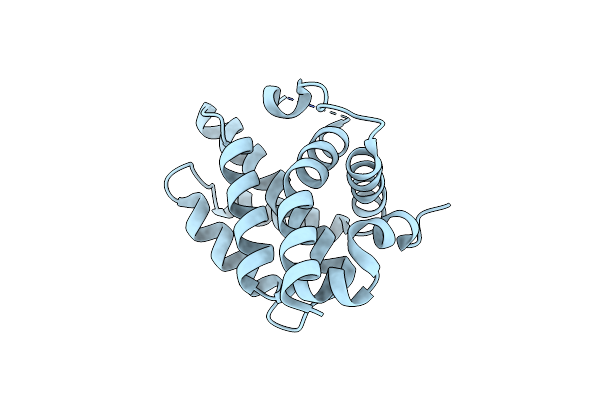 |
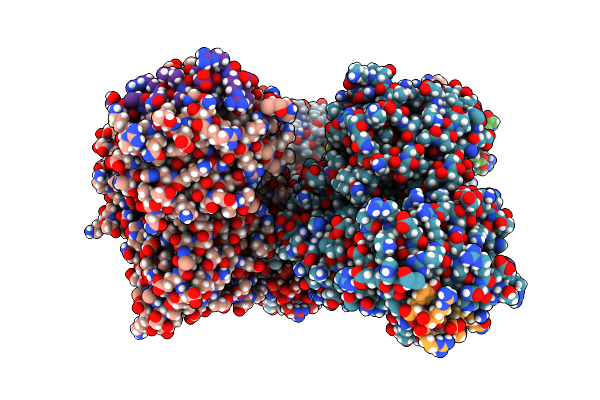
Structural Basis Of Bak Sequestration By Mcl-1 And Consequences For Apoptosis Initiation
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.49 Å Release Date: 2025-06-04 Classification: APOPTOSIS |
 |
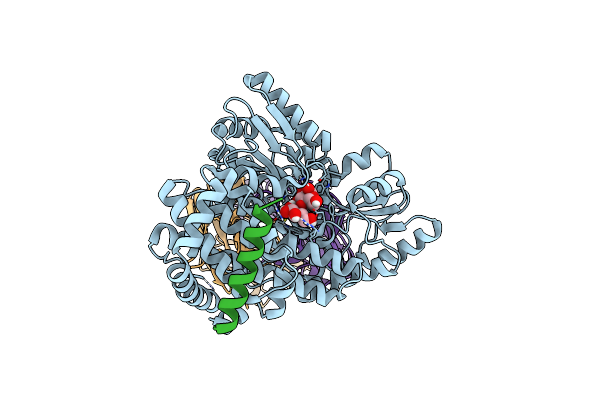
Structural Basis Of Bak Sequestration By Mcl-1 And Consequences For Apoptosis Initiation
Organism: Synthetic construct, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2025-06-04 Classification: APOPTOSIS |
 |
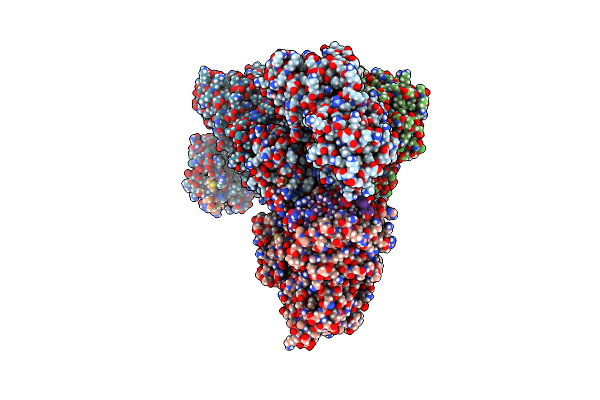
Structural Basis Of Bak Sequestration By Mcl-1 And Consequences For Apoptosis Initiation
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-06-04 Classification: APOPTOSIS |
 |
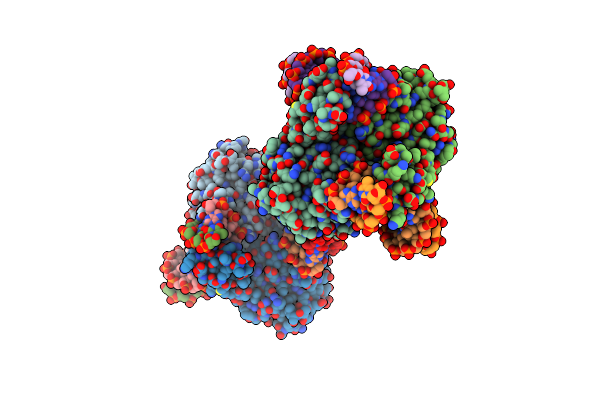
Structural Basis Of Bak Sequestration By Mcl-1 And Consequences For Apoptosis Initiation
Organism: Synthetic construct, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.89 Å Release Date: 2025-06-04 Classification: APOPTOSIS |
 |
Crystal Structure Of Dna N6-Adenine Methyltransferase M.Bcejiv From Burkholderia Cenocepacia In Complex With Duplex Dna Substrates
Organism: Burkholderia cenocepacia
Method: X-RAY DIFFRACTION Resolution:2.11 Å Release Date: 2024-09-11 Classification: DNA BINDING PROTEIN Ligands: EPE, SFG |
 |
Crystal Structure Of Dna N6-Adenine Methyltransferase M.Bcejiv From Burkholderia Cenocepacia In Complex With Duplex Dna Substrate Containing Gtatac As Recognition Sequence
Organism: Burkholderia cenocepacia
Method: X-RAY DIFFRACTION Resolution:2.16 Å Release Date: 2024-09-11 Classification: DNA BINDING PROTEIN Ligands: SFG |
 |
Crystal Structure Of Dna N6-Adenine Methyltransferase M.Bcejiv From Burkholderia Cenocepacia In Complex With Duplex Dna Substrate Containing Gtaaac As Recognition Sequence
Organism: Burkholderia cenocepacia
Method: X-RAY DIFFRACTION Resolution:2.37 Å Release Date: 2024-09-11 Classification: DNA BINDING PROTEIN Ligands: SFG |
 |
Crystal Structure Of Dna N6-Adenine Methyltransferase M.Bcejiv From Burkholderia Cenocepacia In Complex With Duplex Dna Substrate Containing Gtttac As Recognition Sequence
Organism: Burkholderia cenocepacia
Method: X-RAY DIFFRACTION Resolution:2.77 Å Release Date: 2024-09-11 Classification: DNA BINDING PROTEIN Ligands: SFG |
 |
Crystal Structure Of H19 Influenza A Virus Hemagglutinin From A/Lesser Scaup/California/3087/2010
Organism: Influenza a virus (a/lesser scaup/california/3087/2010)
Method: X-RAY DIFFRACTION Resolution:2.38 Å Release Date: 2024-07-03 Classification: VIRAL PROTEIN Ligands: NAG |
 |
Organism: Thermoflavifilum thermophilum, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2024-06-12 Classification: RNA BINDING PROTEIN/RNA/DNA Ligands: MG |
 |
Crystal Structure Of Apo Form Of Short Prokaryotic Argonaute Tir-Apaz (Sparta) Heterodimer
Organism: Thermoflavifilum thermophilum
Method: X-RAY DIFFRACTION Resolution:2.66 Å Release Date: 2024-06-12 Classification: RNA BINDING PROTEIN Ligands: PRO, MN |
 |
Organism: Saccharomyces cerevisiae, Dna molecule
Method: ELECTRON MICROSCOPY Release Date: 2024-05-15 Classification: DNA BINDING PROTEIN/DNA Ligands: CA, DCP, SF4 |
 |
Organism: Saccharomyces cerevisiae, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2024-05-15 Classification: DNA BINDING PROTEIN/DNA Ligands: SF4, CA, DCP |
 |
Structure Of Cbass Cap5 From Pseudomonas Syringae In The Absence Of A Ligand (Apo Form Dimer)
Organism: Pseudomonas syringae
Method: X-RAY DIFFRACTION Resolution:3.16 Å Release Date: 2024-02-07 Classification: IMMUNE SYSTEM Ligands: ZN |
 |
Structure Of Cbass Cap5 From Pseudomonas Syringae As An Activated Tetramer With The Cyclic Dinucleotide 3'2'-C-Diamp Ligand (1 Tetramer In The Au)
Organism: Pseudomonas syringae
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2024-02-07 Classification: IMMUNE SYSTEM Ligands: Y4F, GOL, RWB, ZN, MG, P6G |
 |
Structure Of Cbass Cap5 From Pseudomonas Syringae As An Activated Tetramer With The Cyclic Dinucleotide 3'2'-C-Diamp Ligand (3 Tetramers In The Au)
Organism: Pseudomonas syringae
Method: X-RAY DIFFRACTION Resolution:1.79 Å Release Date: 2024-02-07 Classification: IMMUNE SYSTEM Ligands: ZN, MG, Y4F |
 |
Structure Of Cbass Cap5 From Pseudomonas Syringae As An Activated Tetramer With The Cyclic Dinucleotide 3'2'-C-Dgamp Ligand (2 Tetramers In The Au)
Organism: Pseudomonas syringae
Method: X-RAY DIFFRACTION Resolution:1.87 Å Release Date: 2024-02-07 Classification: IMMUNE SYSTEM Ligands: 4UR, ZN, MG, PGE, PEG, GOL |

