Search Count: 56
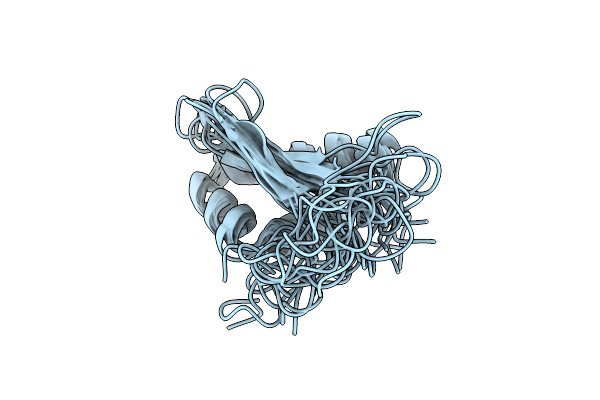 |
Solution Nmr Structure Of The Ripp Proteusin Dpre2 From Desulfotomaculum Sp.
Organism: Desulfotomaculum sp.
Method: SOLUTION NMR Release Date: 2024-12-04 Classification: BIOSYNTHETIC PROTEIN |
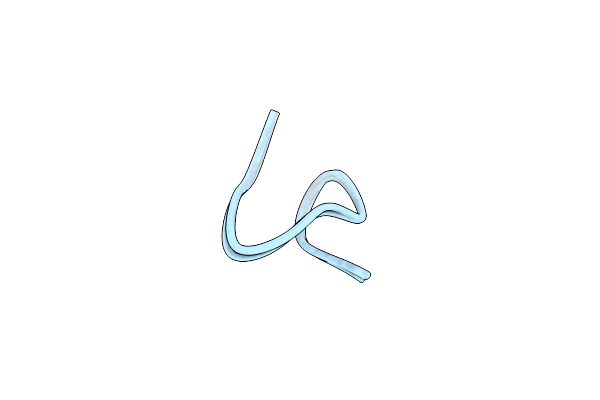 |
Solution Nmr Structure Of Halichondamide A, A Fused Bicyclic Cysteine Knot Undecapeptide From The Marine Sponge Halichondria Bowerbanki
Organism: Halichondria bowerbanki
Method: SOLUTION NMR Release Date: 2024-08-21 Classification: BIOSYNTHETIC PROTEIN |
 |
Organism: Methylovulum psychrotolerans, Synthetic construct
Method: SOLUTION NMR Release Date: 2024-02-14 Classification: BIOSYNTHETIC PROTEIN |
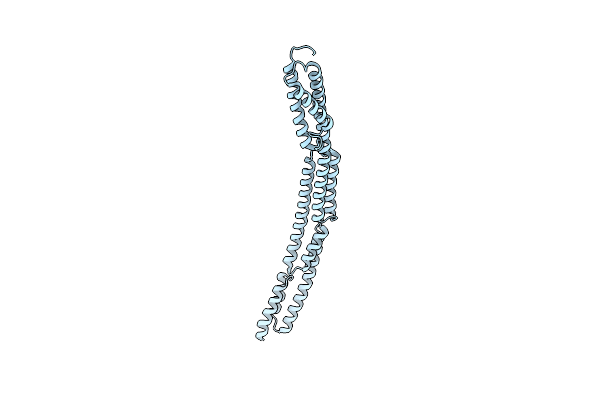 |
Novel Structure Of The N-Terminal Helical Domain Of Biba, A Group B Streptococcus Immunogenic Bacterial Adhesin
Organism: Streptococcus agalactiae
Method: X-RAY DIFFRACTION Resolution:3.03 Å Release Date: 2020-08-12 Classification: IMMUNE SYSTEM |
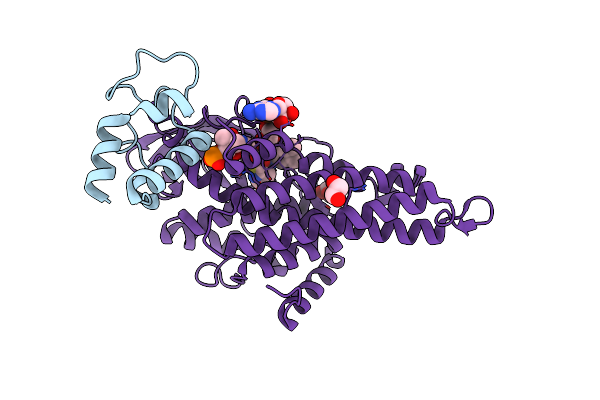 |
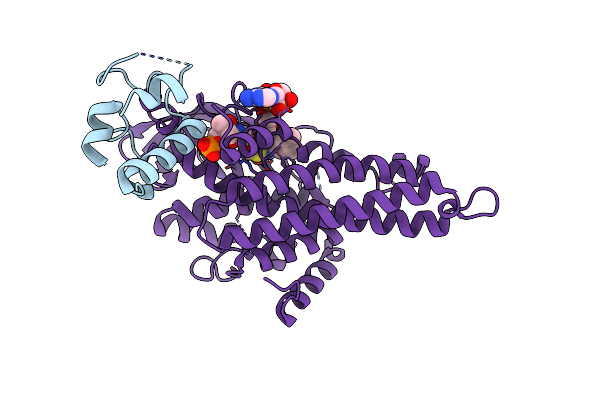
Organism: Marinomonas mediterranea (strain atcc 700492 / jcm 21426 / nbrc 103028 / mmb-1)
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2019-01-16 Classification: OXIDOREDUCTASE Ligands: FK4, FAD, EDO |
 |
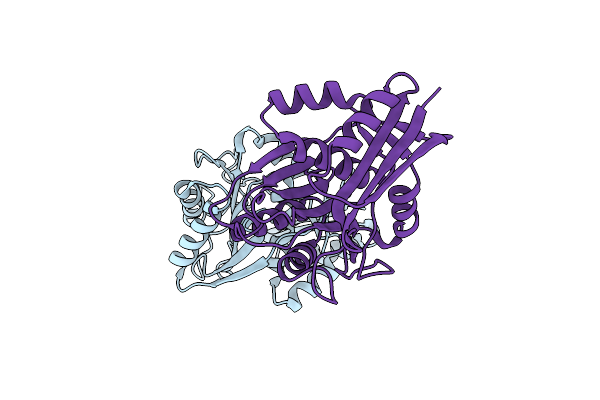
Organism: Marinomonas mediterranea (strain atcc 700492 / jcm 21426 / nbrc 103028 / mmb-1)
Method: X-RAY DIFFRACTION Resolution:2.73 Å Release Date: 2019-01-16 Classification: BIOSYNTHETIC PROTEIN Ligands: FAD, PNS |
 |
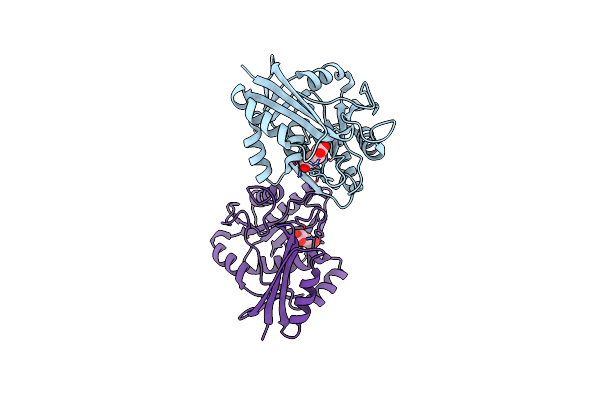
Organism: Aquifex aeolicus
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2016-12-07 Classification: LIGASE |
 |
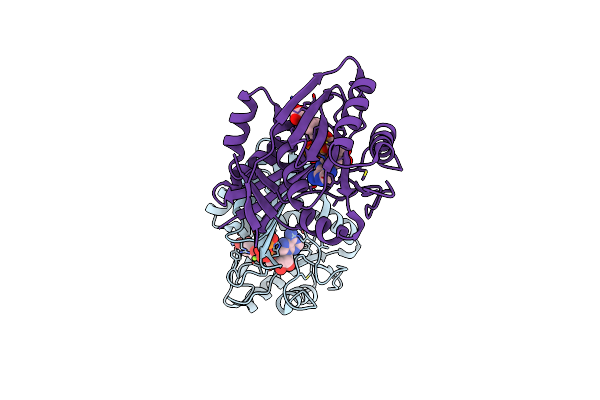
Organism: Aquifex aeolicus
Method: X-RAY DIFFRACTION Resolution:2.46 Å Release Date: 2016-12-07 Classification: LIGASE Ligands: PML |
 |
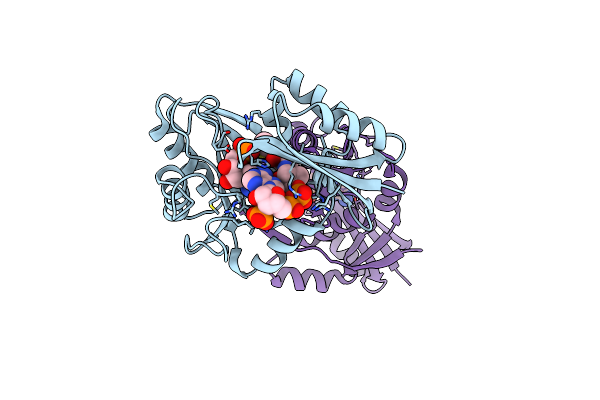
Organism: Aquifex aeolicus
Method: X-RAY DIFFRACTION Resolution:2.55 Å Release Date: 2016-12-07 Classification: LIGASE Ligands: APC, MG, PML |
 |
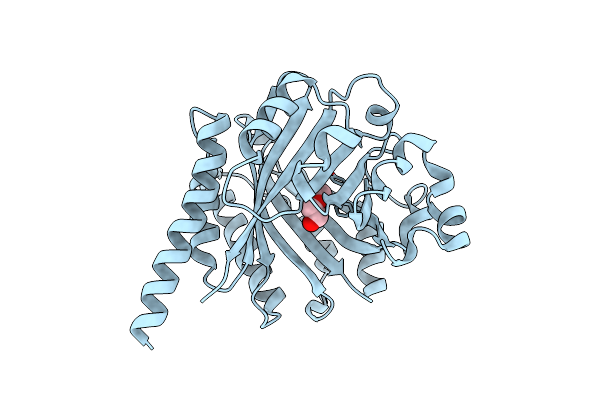
Organism: Aquifex aeolicus
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2016-12-07 Classification: LIGASE Ligands: AMP, COA |
 |
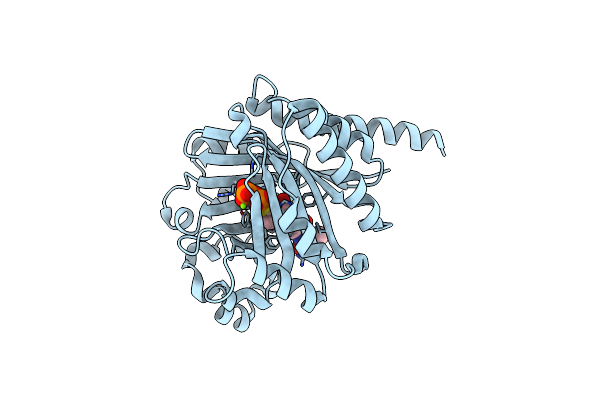
Organism: Planktothrix agardhii nies-596
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2016-11-30 Classification: TRANSFERASE Ligands: PEG |
 |
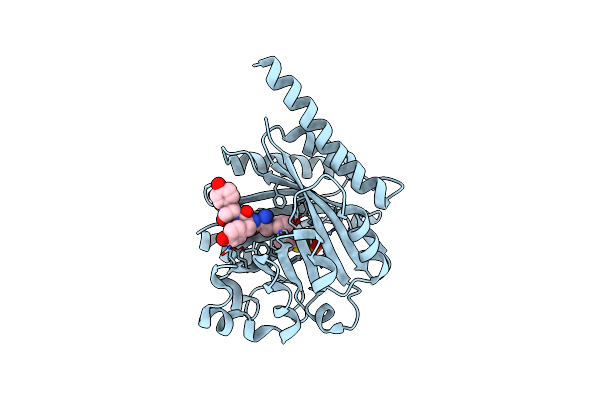
Organism: Planktothrix agardhii nies-596
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2016-11-30 Classification: TRANSFERASE Ligands: DST, MG, B1C |
 |
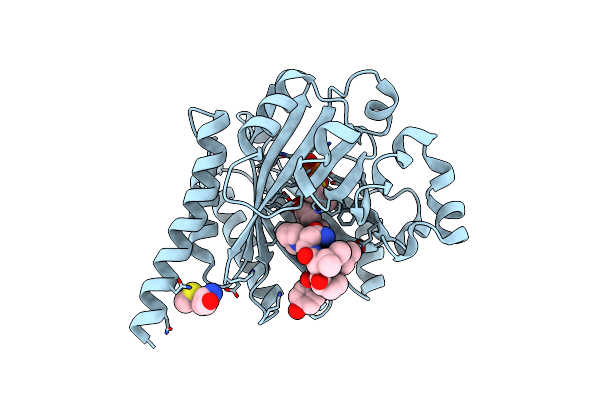
Organism: Planktothrix agardhii nies-596, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2016-11-30 Classification: TRANSFERASE Ligands: DST, MG |
 |
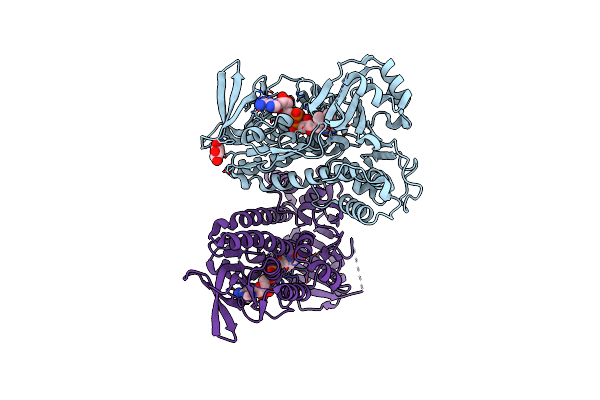
Organism: Planktothrix agardhii nies-596, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.22 Å Release Date: 2016-11-30 Classification: TRANSFERASE Ligands: MG, MET, DST |
 |
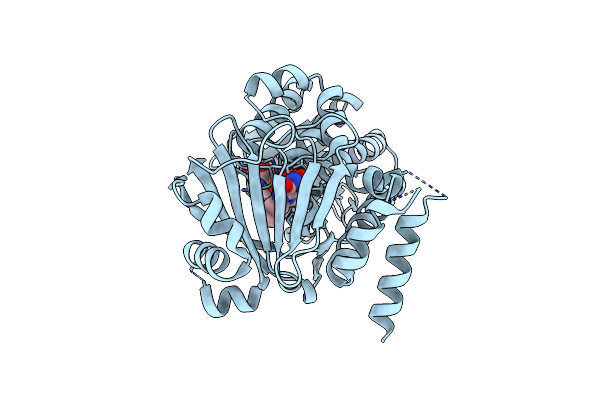
Organism: Streptomyces sp. cnq-418
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2016-03-09 Classification: OXIDOREDUCTASE Ligands: FAD, GOL |
 |
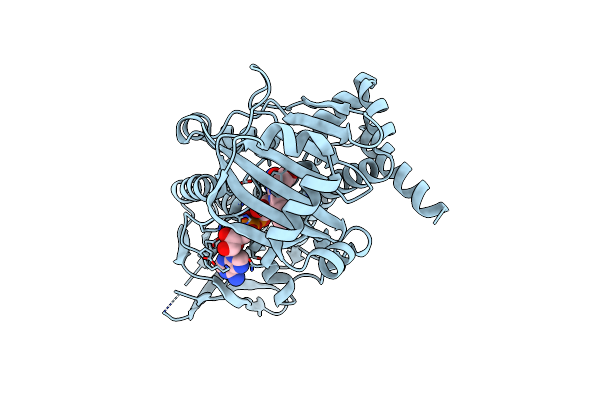
Structure Of Flavin-Dependent Brominase Bmp2 Triple Mutant Y302S F306V A345W
Organism: Streptomyces
Method: X-RAY DIFFRACTION Resolution:1.98 Å Release Date: 2016-03-09 Classification: HYDROLASE Ligands: FAD |
 |
Organism: Streptomyces
Method: X-RAY DIFFRACTION Resolution:1.87 Å Release Date: 2016-03-09 Classification: HYDROLASE Ligands: FAD, EDO |
 |
Organism: Caldanaerobius polysaccharolyticus
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2014-10-29 Classification: SUGAR BINDING PROTEIN |
 |
Organism: Caldanaerobius polysaccharolyticus
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2014-10-29 Classification: SUGAR BINDING PROTEIN Ligands: ZN |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.23 Å Release Date: 2013-12-25 Classification: HYDROLASE Ligands: 1EG, EDO |

