Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
 |
Organism: Bacillus cereus bdrd-cer4
Method: X-RAY DIFFRACTION Resolution:2.26 Å Release Date: 2018-11-14 Classification: MEMBRANE PROTEIN Ligands: K, MPD |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Resolution:4.40 Å Release Date: 2017-09-13 Classification: TRANSCRIPTION Ligands: SF4 |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.79 Å Release Date: 2017-08-16 Classification: CHAPERONE Ligands: IMD, ZN, CL |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.87 Å Release Date: 2017-08-16 Classification: CHAPERONE Ligands: IMD, ZN, CL, GLU |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.77 Å Release Date: 2017-08-16 Classification: CHAPERONE Ligands: IMD, ZN, CL |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.87 Å Release Date: 2017-08-16 Classification: CHAPERONE Ligands: ZN, CL, IMD |
 |
Organism: Salmonella phage p22
Method: ELECTRON MICROSCOPY Release Date: 2017-03-15 Classification: VIRUS |
 |
Organism: Enterobacteria phage p22
Method: X-RAY DIFFRACTION Resolution:3.30 Å Release Date: 2017-02-08 Classification: VIRAL PROTEIN |
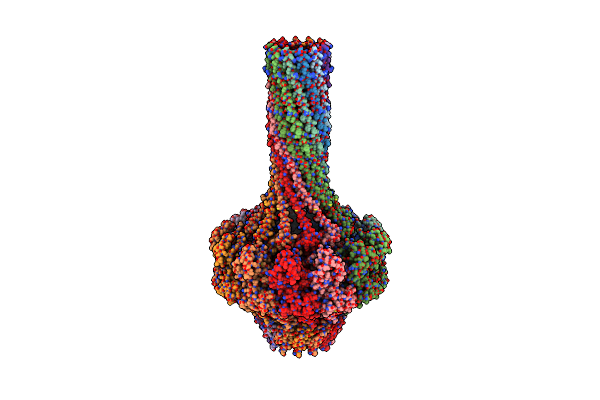 |
Organism: Enterobacteria phage p22
Method: X-RAY DIFFRACTION Resolution:7.00 Å Release Date: 2017-02-08 Classification: VIRAL PROTEIN |
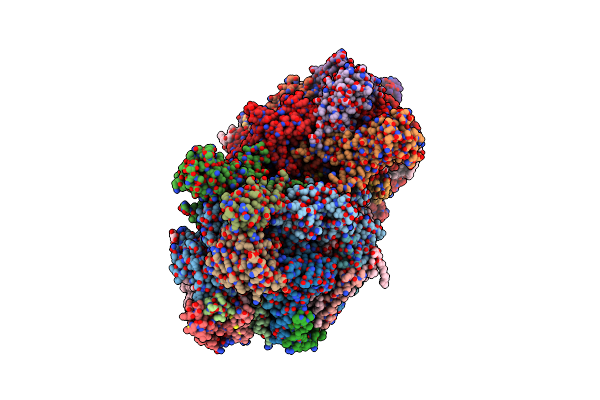 |
Organism: Thermosynechococcus elongatus (strain bp-1)
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2016-11-23 Classification: ELECTRON TRANSPORT Ligands: OEX, FE2, SQD, CL, CLA, PHO, BCR, PL9, LMG, UNL, LHG, BCT, DGD, HEM, HEC |
 |
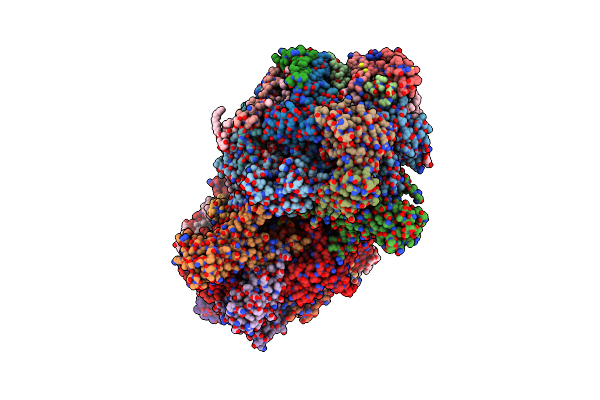
Nh3-Bound Rt Xfel Structure Of Photosystem Ii 500 Ms After The 2Nd Illumination (2F) At 2.8 A Resolution
Organism: Thermosynechococcus elongatus (strain bp-1)
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2016-11-23 Classification: ELECTRON TRANSPORT Ligands: OEX, FE2, LMG, CL, CLA, PHO, BCR, PL9, SQD, UNL, BCT, LHG, DGD, HEM, HEC |
 |
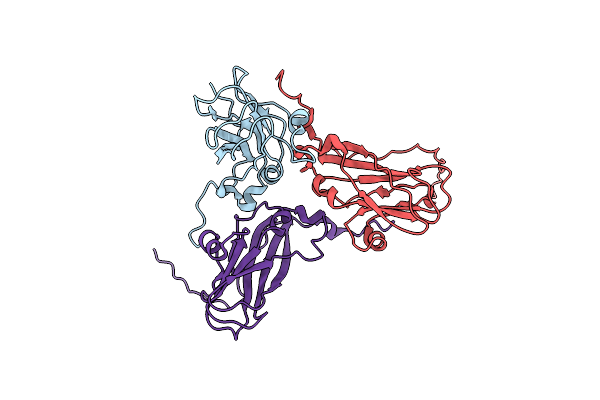
Room Temperature Xfel Structure Of The Native, Doubly-Illuminated Photosystem Ii Complex
Organism: Thermosynechococcus elongatus (strain bp-1), Thermosynechococcus elongatus
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2016-11-23 Classification: PHOTOSYNTHESIS Ligands: OEX, FE2, CL, BCT, CLA, PHO, BCR, PL9, SQD, LHG, UNL, LMG, DGD, HEM, HEC |
 |
Organism: Brome mosaic virus
Method: ELECTRON MICROSCOPY Release Date: 2014-09-10 Classification: VIRUS |
 |
Organism: Brome mosaic virus
Method: ELECTRON MICROSCOPY Release Date: 2014-09-10 Classification: VIRUS |
 |
Organism: Brome mosaic virus
Method: ELECTRON MICROSCOPY Release Date: 2014-09-10 Classification: VIRUS |
 |
Validated Near-Atomic Resolution Structure Of Bacteriophage Epsilon15 Derived From Cryo-Em And Modeling
Organism: Salmonella phage epsilon15
Method: ELECTRON MICROSCOPY Release Date: 2013-07-10 Classification: VIRUS |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.03 Å Release Date: 2010-11-10 Classification: SIGNALING PROTEIN Ligands: KN0 |
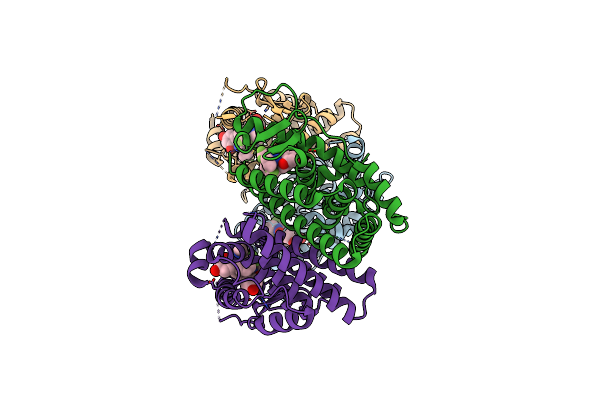 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2010-11-10 Classification: SIGNALING PROTEIN Ligands: KN1 |
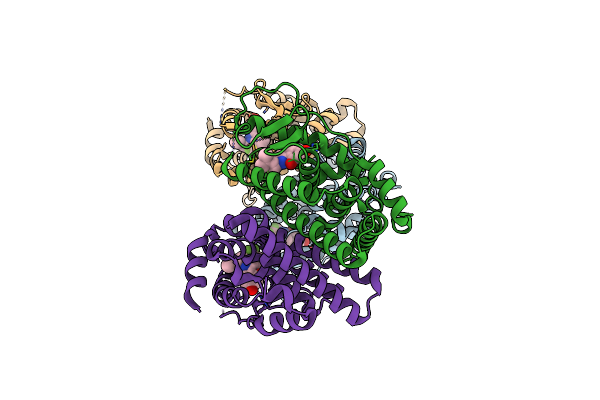 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2010-11-10 Classification: SIGNALING PROTEIN Ligands: KN3 |
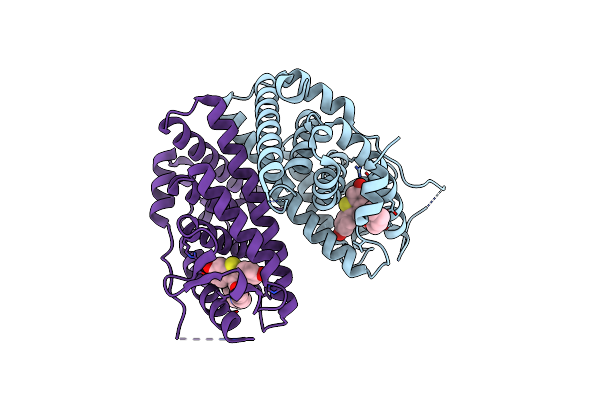 |
Crystal Structure Of Antagonizing Mutant 536S Of The Estrogen Receptor Alpha Ligand Binding Domain Complexed To Raloxifene
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2008-09-02 Classification: TRANSCRIPTION Ligands: RAL |