Search Count: 15
 |
Organism: Thermus phage g20c
Method: X-RAY DIFFRACTION Resolution:2.97 Å Release Date: 2022-11-02 Classification: VIRAL PROTEIN |
 |
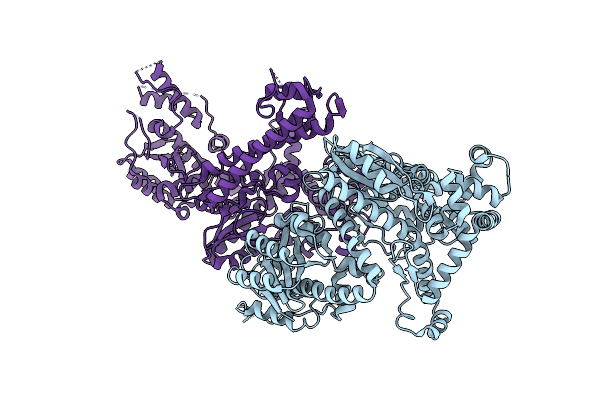
Organism: Thermus phage tsp4
Method: X-RAY DIFFRACTION Resolution:2.19 Å Release Date: 2022-11-02 Classification: VIRAL PROTEIN Ligands: TMP, MG, TB, CL |
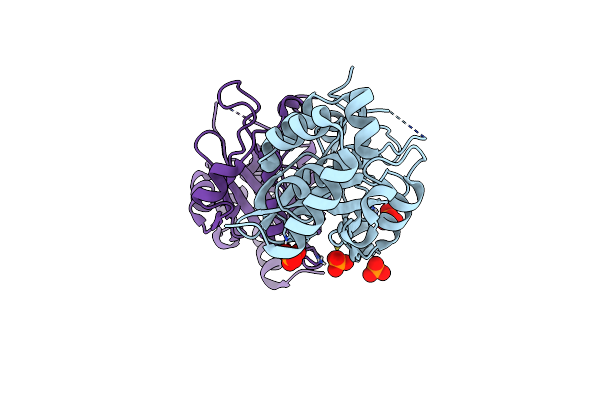 |
Crystal Form 3 Of Cpr-C4: A Cysteine Protease From The Candidate Phyla Radiation
Organism: Candidate division cpr1
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2022-07-06 Classification: HYDROLASE Ligands: PO4 |
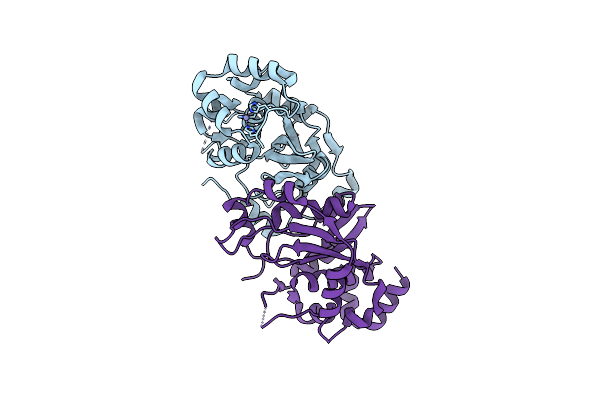 |
Organism: Candidate division cpr1
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2022-04-27 Classification: HYDROLASE Ligands: ZN |
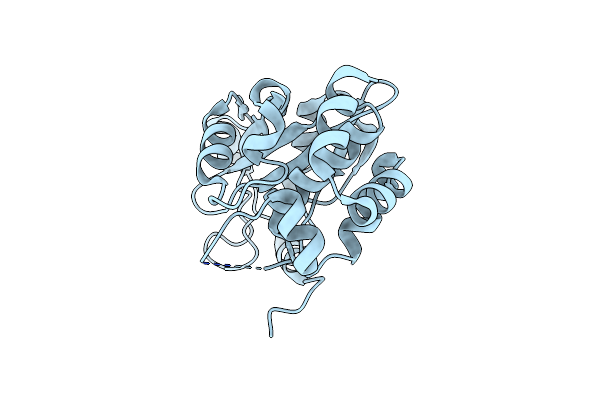 |
Organism: Candidate division cpr1
Method: X-RAY DIFFRACTION Resolution:2.68 Å Release Date: 2022-04-27 Classification: HYDROLASE |
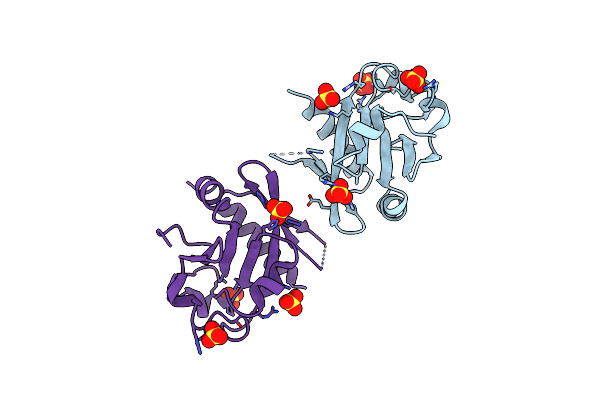 |
Organism: Thermus thermophilus phage 15-6
Method: X-RAY DIFFRACTION Resolution:2.55 Å Release Date: 2022-02-16 Classification: RECOMBINATION Ligands: SO4 |
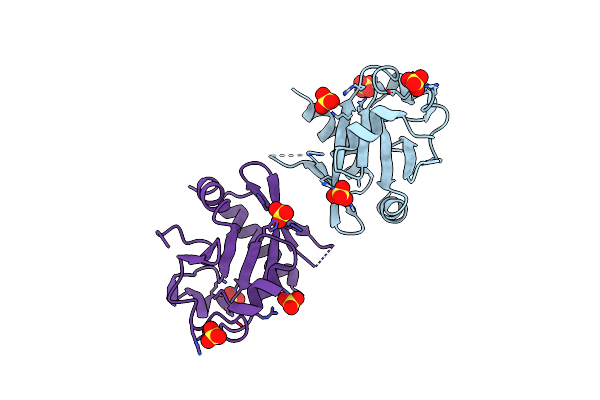 |
Organism: Thermus thermophilus phage 15-6
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2022-01-19 Classification: RECOMBINATION Ligands: SO4 |
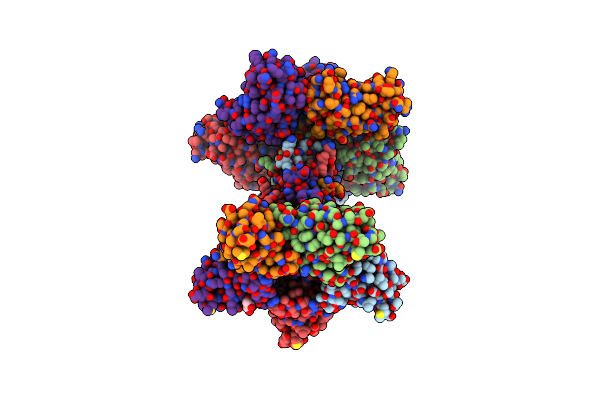 |
Organism: Bacillus subtilis subsp. subtilis str. 168
Method: X-RAY DIFFRACTION Resolution:2.12 Å Release Date: 2019-11-20 Classification: UNKNOWN FUNCTION Ligands: GOL |
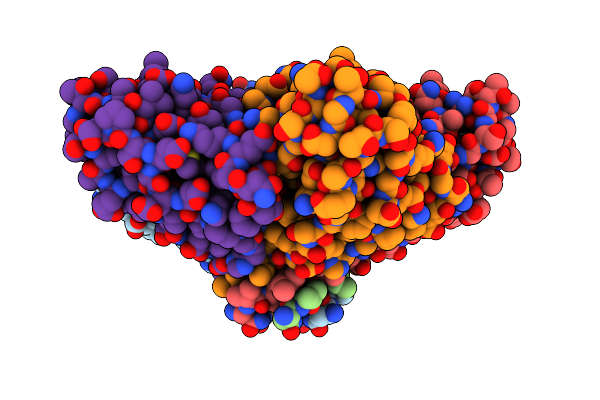 |
Organism: Bacillus subtilis (strain 168)
Method: X-RAY DIFFRACTION Resolution:1.33 Å Release Date: 2019-11-20 Classification: UNKNOWN FUNCTION |
 |
Organism: Bacillus subtilis (strain 168)
Method: X-RAY DIFFRACTION Resolution:1.88 Å Release Date: 2019-11-20 Classification: VIRAL PROTEIN Ligands: GOL, ACT |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2000-03-27 Classification: OXIDOREDUCTASE Ligands: MG, K, TPP |
 |
Crystal Structure Of Pseudomonas Putida 2-Oxoisovalerate Dehydrogenase (Branched-Chain Alpha-Keto Acid Dehydrogenase, E1B)
Organism: Pseudomonas putida
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 1999-08-18 Classification: OXIDOREDUCTASE Ligands: MG, TPP, COI |
 |
Dihydrolipoyl Transacetylase (E.C.2.3.1.12) Catalytic Domain (Residues 184-425) From Bacillus Stearothermophilus
Organism: Geobacillus stearothermophilus
Method: X-RAY DIFFRACTION Resolution:4.40 Å Release Date: 1999-02-16 Classification: ACYLTRANSFERASE |
 |
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 1996-08-01 Classification: ELONGATION FACTOR |
 |
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 1996-07-11 Classification: TRANSLATIONAL GTPASE Ligands: GDP |

