Search Count: 13
 |
Organism: Streptomyces kasugaensis
Method: X-RAY DIFFRACTION Resolution:2.44 Å Release Date: 2022-11-16 Classification: ISOMERASE |
 |
The Crystal Structure Of Non-Hydrolyzing Udpglcnac 2-Epimerase In Complex With Udp-Glucose
Organism: Streptomyces kasugaensis
Method: X-RAY DIFFRACTION Resolution:2.09 Å Release Date: 2022-11-16 Classification: ISOMERASE Ligands: UPG, NA |
 |
The Crystal Structure Of Non-Hydrolyzing Udpglcnac 2-Epimerase In Complex With Udp
Organism: Streptomyces kasugaensis
Method: X-RAY DIFFRACTION Resolution:2.58 Å Release Date: 2022-11-16 Classification: ISOMERASE Ligands: UDP, NA |
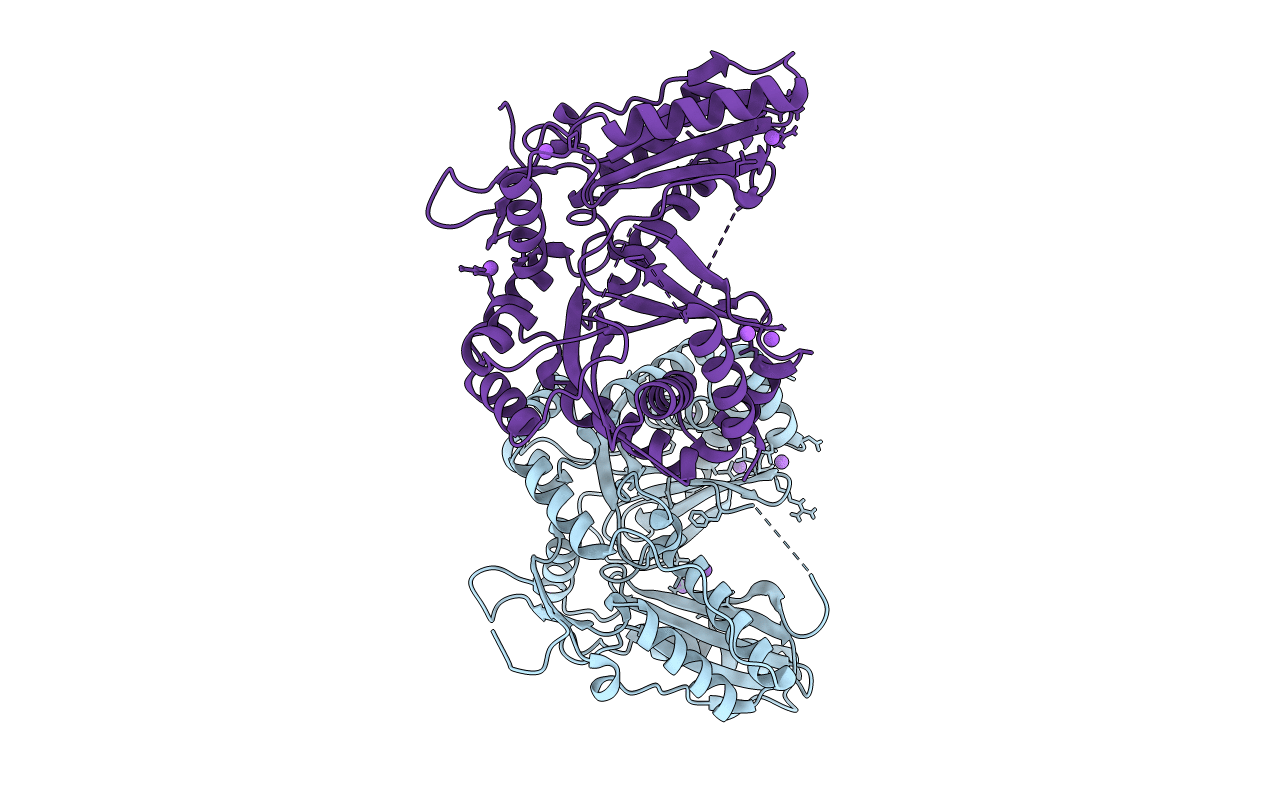 |
Crystal Structure Of A Dehydrating Condensation Domain, Ambe-Cmodaa, Involved In Nonribosomal Peptide Synthesis
Organism: Pseudomonas aeruginosa (strain atcc 15692 / dsm 22644 / cip 104116 / jcm 14847 / lmg 12228 / 1c / prs 101 / pao1)
Method: X-RAY DIFFRACTION Resolution:2.14 Å Release Date: 2022-07-13 Classification: BIOSYNTHETIC PROTEIN Ligands: IOD, NA |
 |
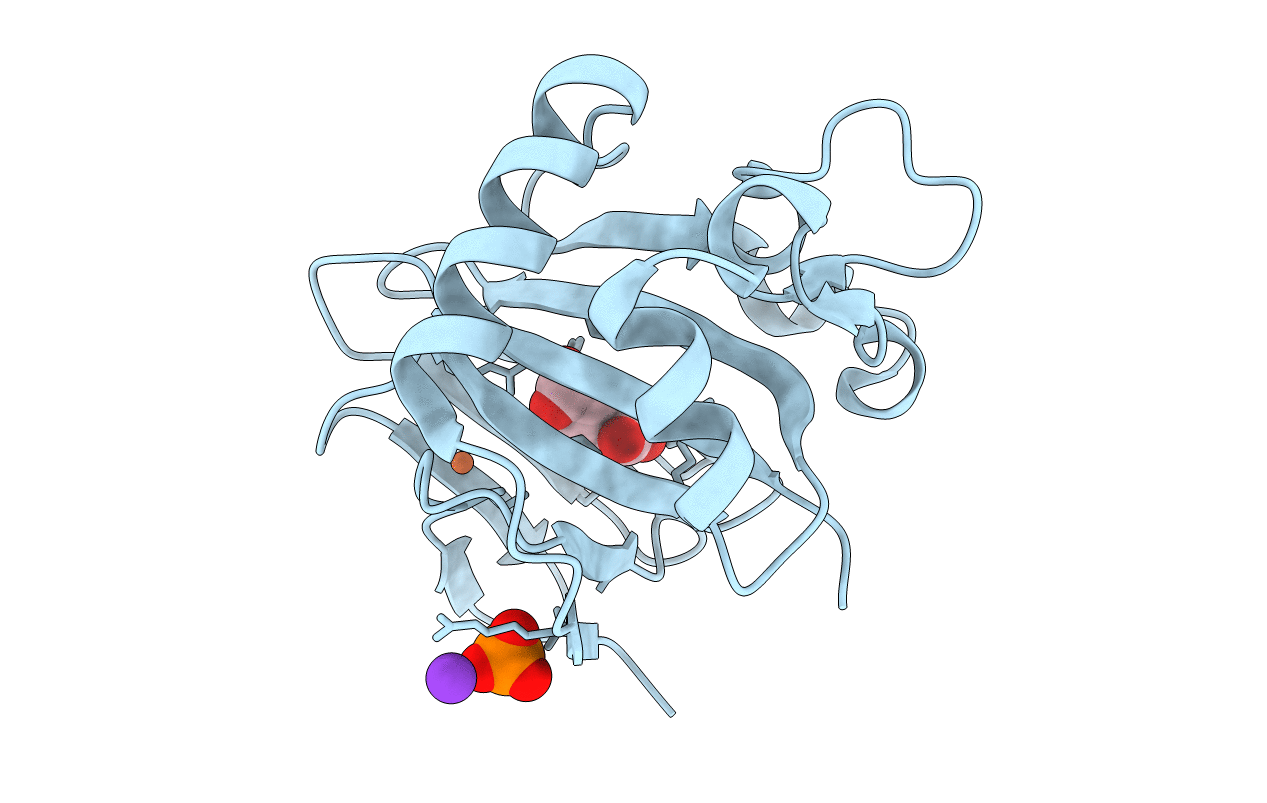
Unique Non-Heme Hydroxylase In Biosynthesis Of Nucleoside Antibiotic Pathway Uncover Mechanism Of Reaction
Organism: Streptomyces sp. mk730-62f2
Method: X-RAY DIFFRACTION Resolution:1.98 Å Release Date: 2022-02-23 Classification: ANTIBIOTIC Ligands: 5KC, SIN, CO2, FE |
 |
Unique Non-Heme Hydroxylase In Biosynthesis Of Nucleoside Antibiotic Pathway Uncover Mechanism Of Reaction
Organism: Streptomyces sp. mk730-62f2
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2022-02-23 Classification: ANTIBIOTIC Ligands: AKG, K, FE, PO4 |
 |
Unique Non-Heme Hydroxylase In Biosynthesis Of Nucleoside Antibiotic Pathway Uncover Mechanism Of Reaction
Organism: Streptomyces sp. mk730-62f2
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2022-02-23 Classification: ANTIBIOTIC Ligands: FE |
 |
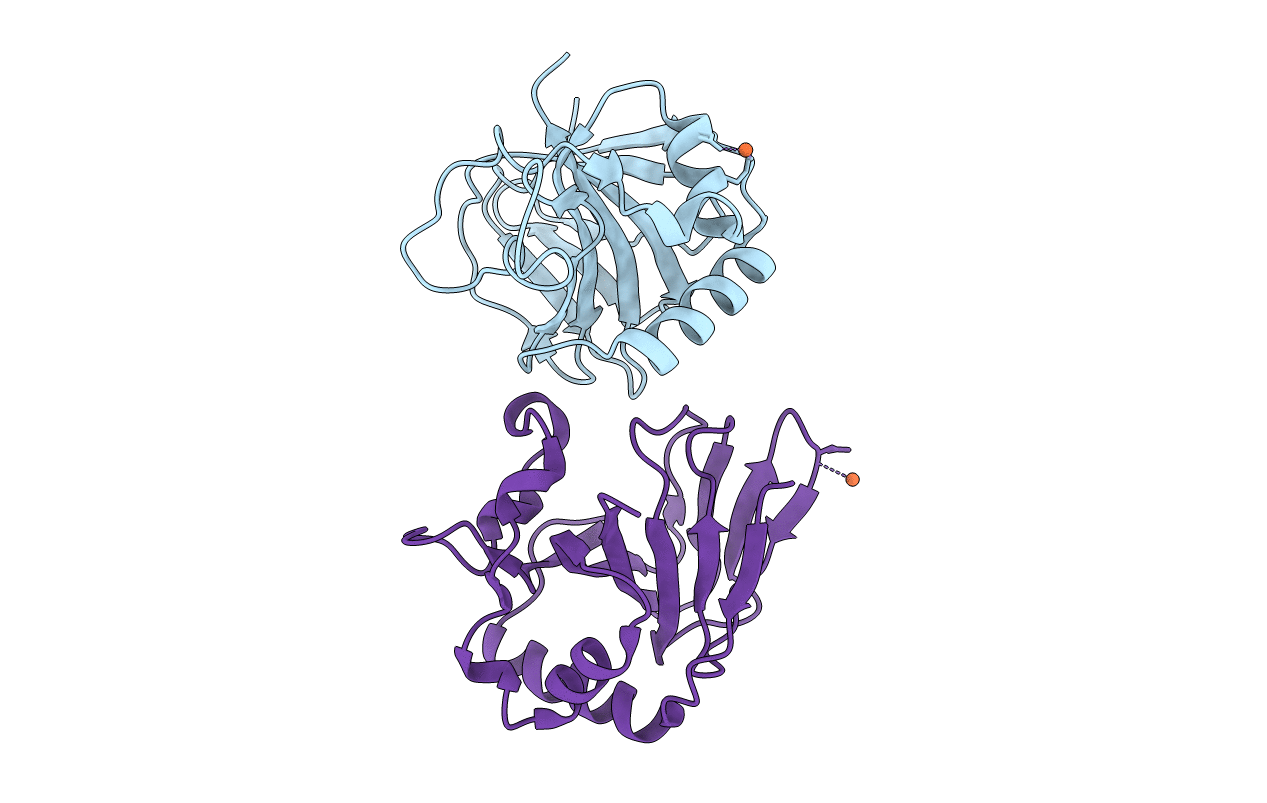
Unique Non-Heme Hydroxylase In Biosynthesis Of Nucleoside Antibiotic Pathway Uncover Mechanism Of Reaction
Organism: Streptomyces sp. mk730-62f2
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2022-02-23 Classification: ANTIBIOTIC Ligands: P3G |
 |
Unique Non-Heme Hydroxylase In Biosynthesis Of Nucleoside Antibiotic Pathway Uncover Mechanism Of Reaction
Organism: Streptomyces sp. mk730-62f2
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2022-02-23 Classification: ANTIBIOTIC Ligands: FE |
 |
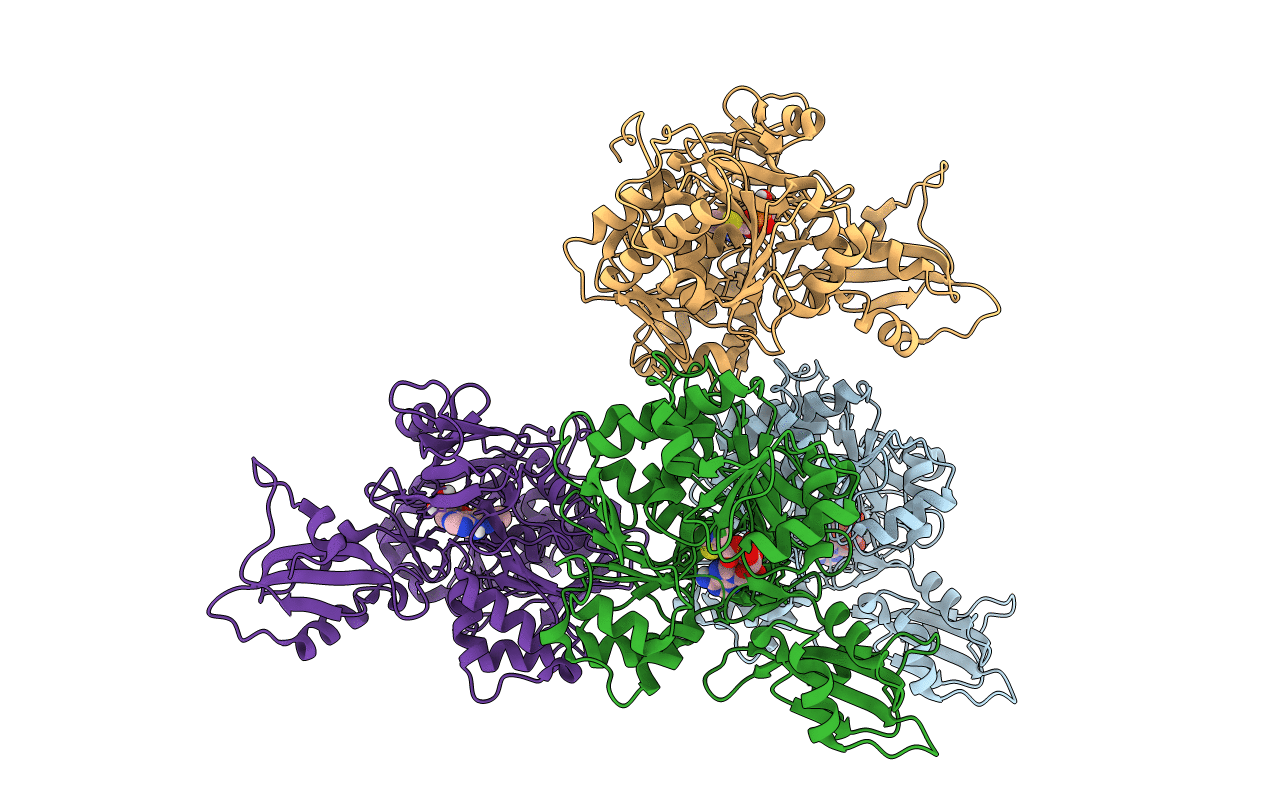
Organism: Aloe arborescens
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2022-01-19 Classification: TRANSFERASE |
 |
The Crystal Structure Of Benzoate Coenzyme A Ligase Double Mutant (H333A/I334A) In Complex With 2-Methyl-Thiazole-5 Carboxylate-Amp
Organism: Rhodopseudomonas palustris
Method: X-RAY DIFFRACTION Resolution:2.14 Å Release Date: 2021-03-03 Classification: LIGASE Ligands: AMP, 6V9 |
 |
Organism: Rhodopseudomonas palustris
Method: X-RAY DIFFRACTION Resolution:1.57 Å Release Date: 2021-01-13 Classification: LIGASE |
 |
The Crystal Structure Of Benzoate Coenzyme A Ligase Double Mutant (H333A/I334A) In Complex With 2-Chloro-1,3-Thiazole-5-Carboxylate-Amp
Organism: Rhodopseudomonas palustris
Method: X-RAY DIFFRACTION Resolution:1.71 Å Release Date: 2021-01-13 Classification: LIGASE Ligands: AMP, F0O |

