Search Count: 16
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.91 Å Release Date: 2016-06-29 Classification: PROTEIN TRANSPORT |
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.83 Å Release Date: 2016-06-29 Classification: HYDROLASE Ligands: ZN |
 |
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2016-06-29 Classification: HYDROLASE Ligands: ZN, CAC |
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:3.40 Å Release Date: 2016-06-29 Classification: HYDROLASE |
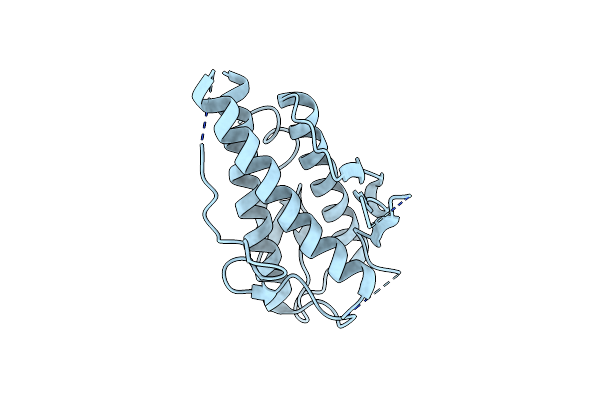 |
Crystal Structure Of The Beta-Alpha Repeated, Autophagy-Specific (Bara) Domain Of Vps30/Atg6
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2012-03-14 Classification: PROTEIN TRANSPORT |
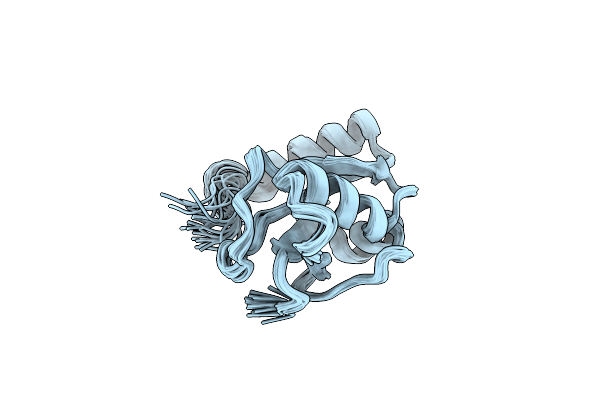 |
Organism: Saccharomyces cerevisiae
Method: SOLUTION NMR Release Date: 2010-05-12 Classification: PROTEIN TRANSPORT |
 |
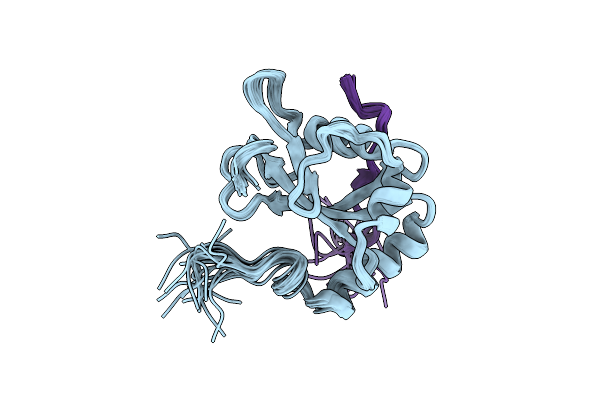
The Crystal Structure Of Saccharomyces Cerevisiae Atg8- Atg19(412-415) Complex
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2008-12-09 Classification: PROTEIN TRANSPORT Ligands: SO4 |
 |
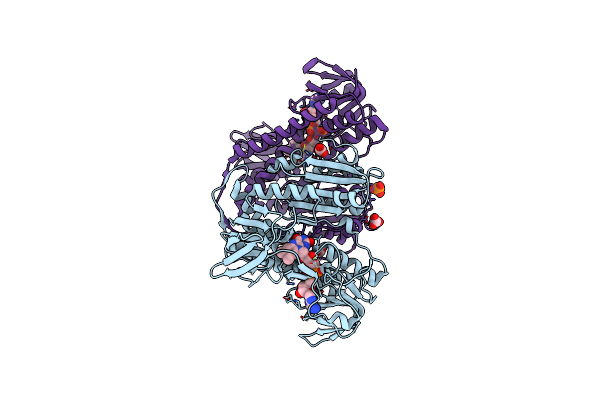
Organism: Rattus norvegicus
Method: SOLUTION NMR Release Date: 2008-09-02 Classification: APOPTOSIS INHIBITOR/APOPTOSIS |
 |
Crystal Structures And Evolutionary Relationship Of Two Different Lipoamide Dehydrogenase(E3S) From Thermus Thermophilus
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2008-04-01 Classification: OXIDOREDUCTASE Ligands: CO3, FAD, PO4 |
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2005-02-15 Classification: OXIDOREDUCTASE Ligands: PO4, FAD, NAD |
 |
The Structure Of The Mutant, S225A And E251L, Of 3-Isopropylmalate Dehydrogenase From Bacillus Coagulans
Organism: Bacillus coagulans
Method: X-RAY DIFFRACTION Resolution:2.95 Å Release Date: 2005-02-15 Classification: OXIDOREDUCTASE Ligands: SO4 |
 |
The Crystal Structure Of The Inactive Form Chitosanase From Bacillus Sp. K17 At Ph3.7
Organism: Bacillus sp.
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2004-12-07 Classification: HYDROLASE Ligands: SO4 |
 |
The Crystal Structure Of The Active Form Chitosanase From Bacillus Sp. K17 At Ph6.4
Organism: Bacillus sp.
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2004-12-07 Classification: HYDROLASE Ligands: PIN |
 |
Crystal Structure Of D(Gcgagagc): The Dna Quadruplex Structure Split From The Octaplex
|
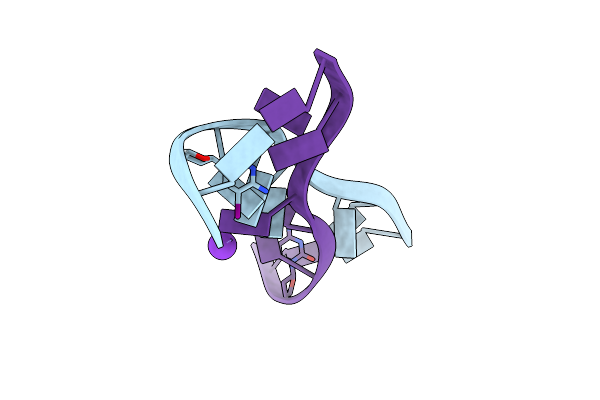 |
Crystal Structure Of D(Gcgagagc): The Dna Quadruplex Structure Split From The Octaplex
|
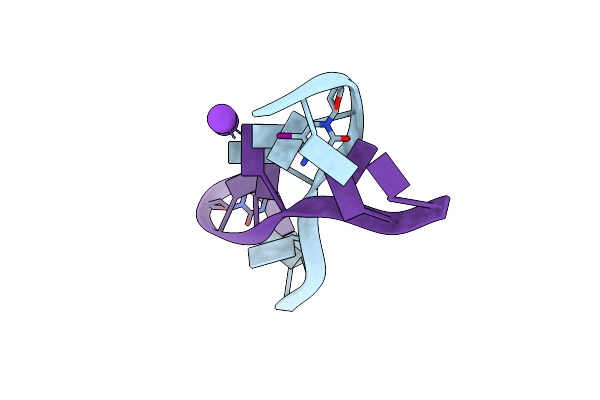 |
Crystal Structure Of D(Gcgagagc): The Dna Octaplex Structure With I-Motif Of G-Quartet
|

