Search Count: 90
 |
Organism: Streptomyces avidinii
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2025-06-04 Classification: UNKNOWN FUNCTION |
 |
Organism: Streptomyces avidinii
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2025-06-04 Classification: UNKNOWN FUNCTION |
 |
Crystal Structure Of N-Terminal Domain Of N-Methyl-D-Aspartate Receptor Subunit Nr1 In Complex With Patient-Derived Antibody
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-05-14 Classification: MEMBRANE PROTEIN |
 |
Organism: Discosoma sp.
Method: X-RAY DIFFRACTION Resolution:1.01 Å Release Date: 2023-09-27 Classification: FLUORESCENT PROTEIN |
 |
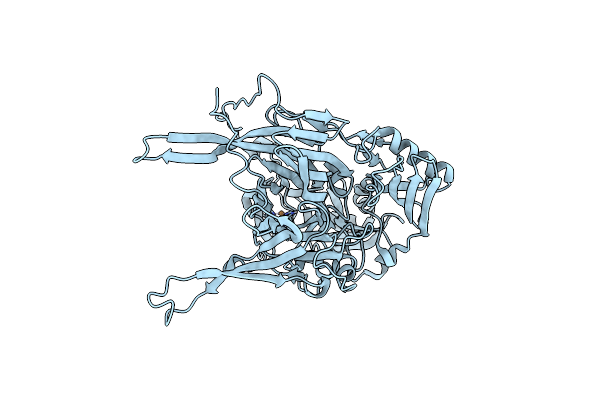
Neutron Structure Of Copper Amine Oxidase From Arthrobacter Globiformis Anaerobically Reduced By Phenylethylamine At Pd 9.0
Organism: Arthrobacter globiformis
Method: X-RAY DIFFRACTION, NEUTRON DIFFRACTION Resolution:1.09 Å, 1.67 Å Release Date: 2023-09-20 Classification: OXIDOREDUCTASE Ligands: CU, NA, PEA |
 |
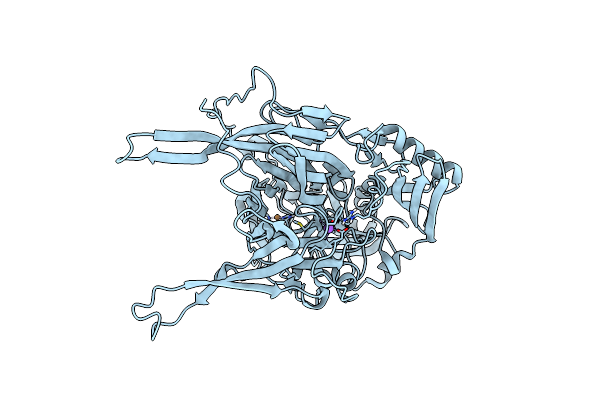
Crystallographic Structure Of Copper Amine Oxidase From Arthrobacter Glibiformis At Pd 7.4 Determined By Only Neutron Diffraction Data.
Organism: Arthrobacter globiformis
Method: NEUTRON DIFFRACTION Resolution:1.72 Å Release Date: 2022-04-20 Classification: OXIDOREDUCTASE Ligands: CU, DOD |
 |
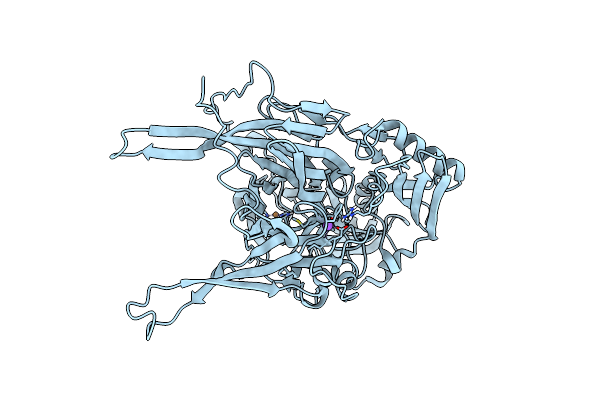
Crystallographic Structure Of Copper Amine Oxidase From Arthrobacter Glibiformis At Pd 7.4 Determined By Both X-Ray And Neutron Diffraction Data At 1.72 Angstrom Resolution.
Organism: Arthrobacter globiformis
Method: X-RAY DIFFRACTION, NEUTRON DIFFRACTION Resolution:1.7200 Å, 1.7200 Å Release Date: 2022-04-20 Classification: OXIDOREDUCTASE Ligands: CU, NA, DOD |
 |
Neutron Structure Of Copper Amine Oxidase From Arthrobacter Glibiformis At Pd 7.4
Organism: Arthrobacter globiformis
Method: X-RAY DIFFRACTION, NEUTRON DIFFRACTION Resolution:1.1400 Å, 1.7200 Å Release Date: 2020-04-29 Classification: OXIDOREDUCTASE Ligands: CU, NA |
 |
Organism: Aequorea victoria
Method: NEUTRON DIFFRACTION Resolution:1.44 Å Release Date: 2020-04-08 Classification: FLUORESCENT PROTEIN Ligands: D3O, DOD |
 |
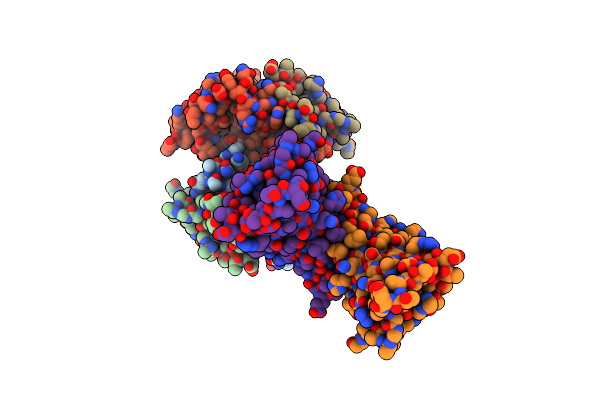
Organism: Aequorea victoria
Method: X-RAY DIFFRACTION Resolution:0.77 Å Release Date: 2020-04-01 Classification: FLUORESCENT PROTEIN Ligands: DOD |
 |
Organism: Streptomyces avidinii
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2020-03-18 Classification: PEPTIDE BINDING PROTEIN Ligands: GOL |
 |
Organism: Halobacterium salinarum (strain atcc 700922 / jcm 11081 / nrc-1)
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2020-01-15 Classification: OXIDOREDUCTASE |
 |
Organism: Human immunodeficiency virus 1
Method: X-RAY DIFFRACTION Resolution:1.00 Å Release Date: 2019-05-22 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: GOL, SO4, B0F, CL |
 |
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2019-02-13 Classification: IMMUNE SYSTEM Ligands: CL |
 |
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2019-02-13 Classification: IMMUNE SYSTEM |
 |
Organism: Deinococcus radiodurans r1
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2018-12-26 Classification: DNA BINDING PROTEIN Ligands: SO4, GOL |
 |
Organism: Deinococcus radiodurans
Method: X-RAY DIFFRACTION Resolution:2.19 Å Release Date: 2018-12-26 Classification: DNA BINDING PROTEIN Ligands: SO4 |
 |
Organism: Deinococcus radiodurans r1
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2018-12-26 Classification: DNA BINDING PROTEIN |
 |
Organism: Leuconostoc mesenteroides subsp. sake
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2018-11-28 Classification: ISOMERASE Ligands: PLP |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION, NEUTRON DIFFRACTION Resolution:1.1000 Å, 1.9000 Å Release Date: 2018-11-21 Classification: TRANSFERASE Ligands: SO4, DOD |

