Search Count: 21
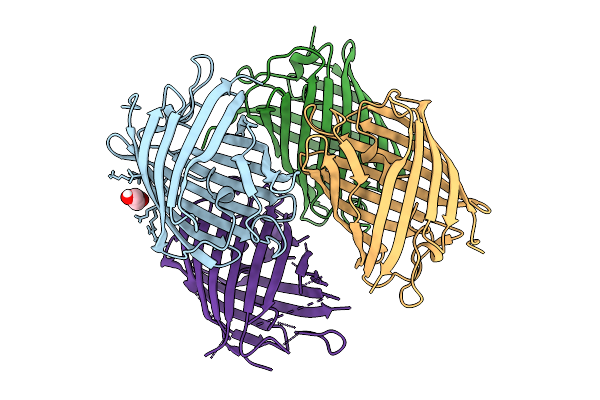 |
X-Ray Crystal Structure Of Kohinoor Reversibly Switchable Fluorescent Protein
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-09-03 Classification: FLUORESCENT PROTEIN Ligands: PEG |
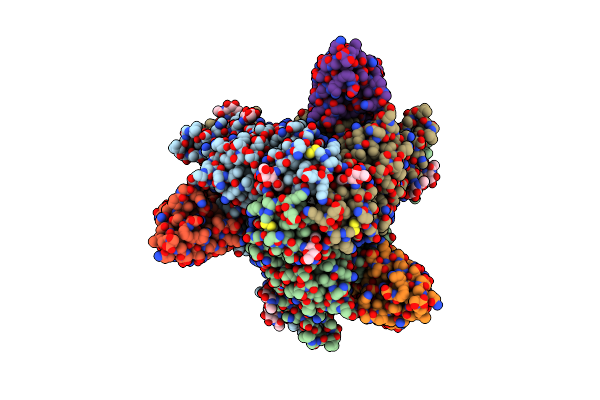 |
Prefusion F Glycoprotein Ectodomain Of Nipah Virus Ectodomain In Complex With Ds90 Nanobody
Organism: Henipavirus nipahense, Vicugna pacos
Method: ELECTRON MICROSCOPY Release Date: 2025-06-04 Classification: VIRAL PROTEIN Ligands: NAG |
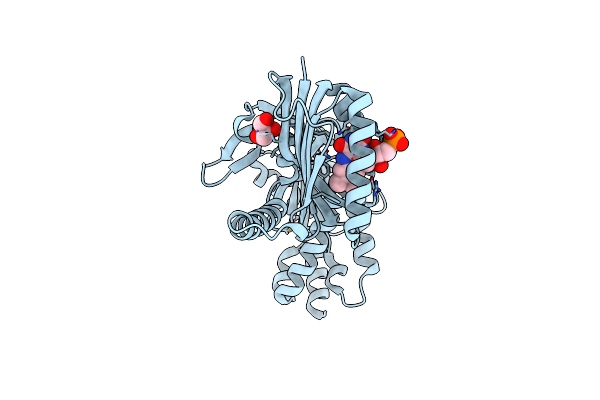 |
Organism: Oscillatoria acuminata
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2025-02-26 Classification: LYASE Ligands: FMN, GOL, CA |
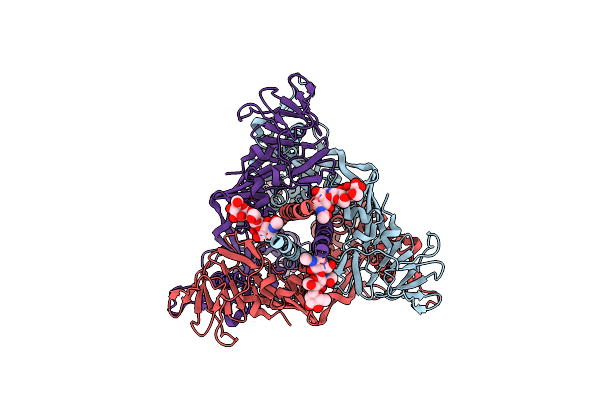 |
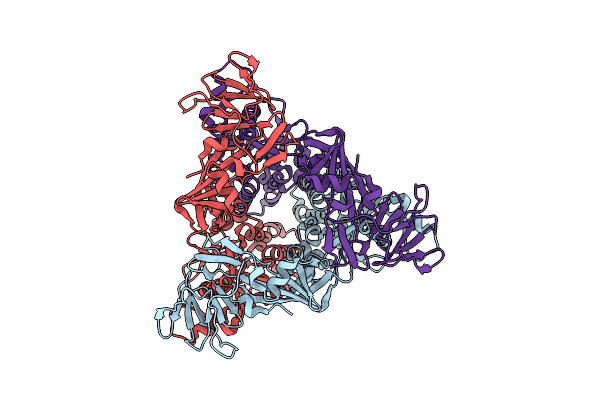
Organism: Langya virus
Method: ELECTRON MICROSCOPY Release Date: 2023-06-28 Classification: VIRAL PROTEIN Ligands: NAG |
 |
Organism: Mojiang virus
Method: ELECTRON MICROSCOPY Release Date: 2023-06-28 Classification: VIRAL PROTEIN |
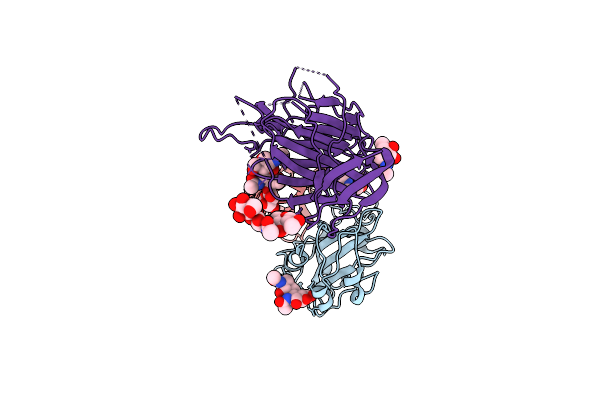 |
Cryo-Em Structure Of Sars-Cov-2 Spike (Hexapro Variant) In Complex With Nanobody W25 (Map 3, Focus Refinement On Rbd, W25 And Adjacent Ntd)
Organism: Severe acute respiratory syndrome coronavirus 2, Vicugna pacos
Method: ELECTRON MICROSCOPY Release Date: 2023-03-08 Classification: VIRAL PROTEIN Ligands: NAG |
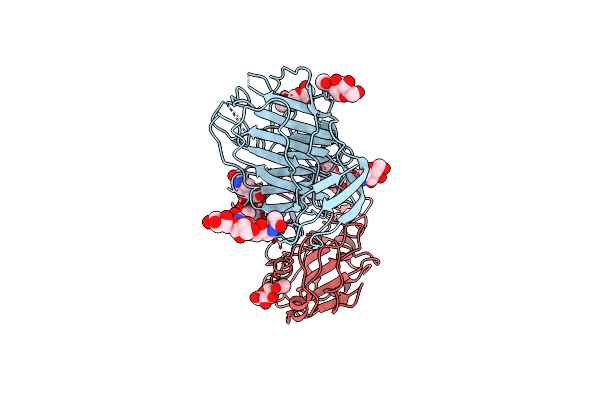 |
Cryo-Em Structure Of Sars-Cov-2 Spike (Omicron Ba.1 Variant) In Complex With Nanobody W25 (Map 5, Focus Refinement On Rbd, W25 And Adjacent Ntd)
Organism: Severe acute respiratory syndrome coronavirus 2, Vicugna pacos
Method: ELECTRON MICROSCOPY Release Date: 2023-03-08 Classification: VIRAL PROTEIN Ligands: NAG |
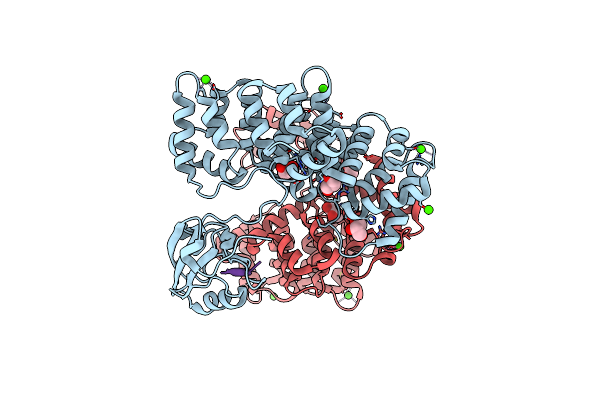 |
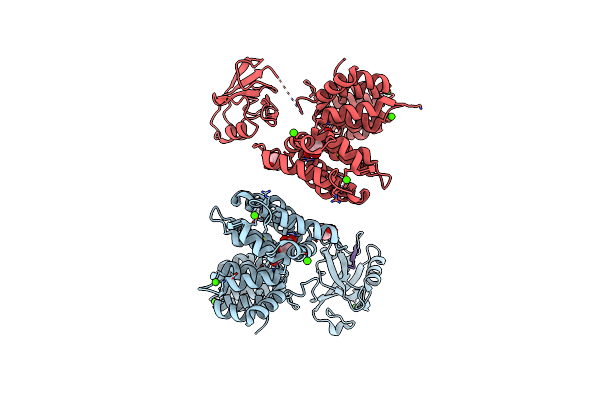
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2017-11-08 Classification: SIGNALING PROTEIN Ligands: GOL, CA |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2017-11-08 Classification: SIGNALING PROTEIN Ligands: GOL, CA |
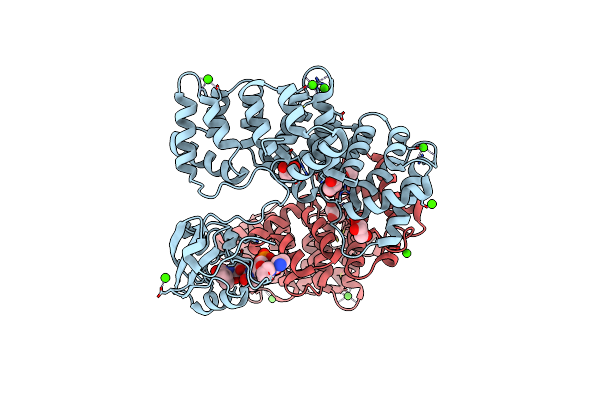 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.95 Å Release Date: 2017-11-08 Classification: SIGNALING PROTEIN Ligands: GOL, CA |
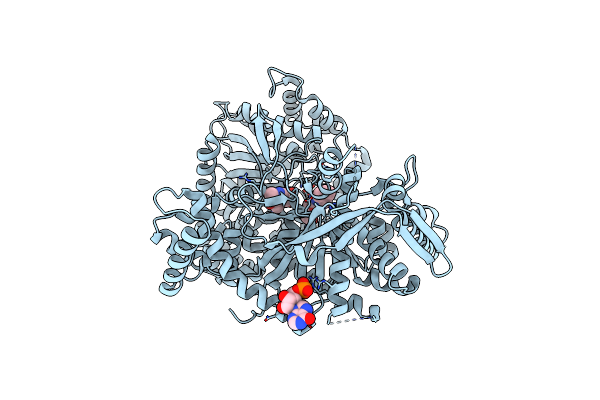 |
Organism: Oryctolagus cuniculus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2017-09-27 Classification: TRANSFERASE Ligands: PLP, 9L2, IMP |
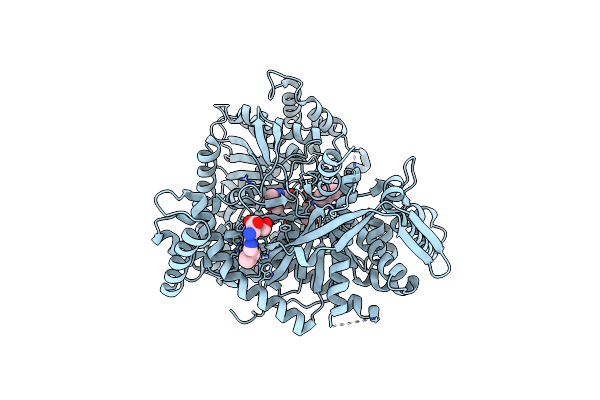 |
Organism: Oryctolagus cuniculus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2017-09-27 Classification: TRANSFERASE Ligands: PLP, 9LE |
 |
Organism: Oryctolagus cuniculus
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2017-09-27 Classification: TRANSFERASE Ligands: PLP, 9LB, IMP |
 |
Organism: Oryctolagus cuniculus
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2017-09-27 Classification: TRANSFERASE Ligands: PLP, 9L8 |
 |
Organism: Oryctolagus cuniculus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2017-05-31 Classification: TRANSFERASE Ligands: KS3, PLP, DMS |
 |
Organism: Oryctolagus cuniculus
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2016-08-24 Classification: TRANSFERASE Ligands: PLP, 6MY, DMS |
 |
Organism: Oryctolagus cuniculus
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2016-08-24 Classification: TRANSFERASE Ligands: PLP, 6NE, DMS |
 |
Crystal Structure Of Bacillus Subtilis Ymdb, A Global Regulator Of Late Adaptive Responses.
Organism: Bacillus subtilis subsp. subtilis
Method: X-RAY DIFFRACTION Resolution:1.64 Å Release Date: 2013-07-24 Classification: HYDROLASE Ligands: PO4, FE2 |
 |
Crystal Structure Of Padr-Like Transcriptional Regulator (Bc4206) From Bacillus Cereus Strain Atcc 14579
Organism: Bacillus cereus
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2012-12-05 Classification: TRANSCRIPTION Ligands: SO4, GOL |
 |
Crystal Structure Of Padr-Like Transcriptional Regulator (Bce3449) From Bacillus Cereus Strain Atcc 10987
Organism: Bacillus cereus
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2012-12-05 Classification: TRANSCRIPTION |

