Search Count: 78
All
Selected
 |
Structure Of The Acinetobacter Calcoaceticus Cyclohexanone Monooxygenase Mutant M10-R327K-R490E-I491E-Y246F-S186W-T187V
Organism: Acinetobacter calcoaceticus
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2025-09-03 Classification: OXIDOREDUCTASE Ligands: FAD |
 |
Structure Of Cyclohexanone Monooxygenase Mutant From Acinetobacter Calcoaceticus
Organism: Acinetobacter calcoaceticus
Method: X-RAY DIFFRACTION Resolution:1.78 Å Release Date: 2025-04-02 Classification: OXIDOREDUCTASE Ligands: FAD, NDP |
 |
Structure Of Cyclohexanone Monooxygenase Mutant From Acinetobacter Calcoaceticus
Organism: Acinetobacter calcoaceticus
Method: X-RAY DIFFRACTION Resolution:2.14 Å Release Date: 2025-03-26 Classification: OXIDOREDUCTASE Ligands: FAD, NDP |
 |
Structure Of Cyclohexanone Monooxygenase Mutant From Acinetobacter Calcoaceticus
Organism: Acinetobacter calcoaceticus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2025-03-26 Classification: OXIDOREDUCTASE Ligands: NDP, FAD |
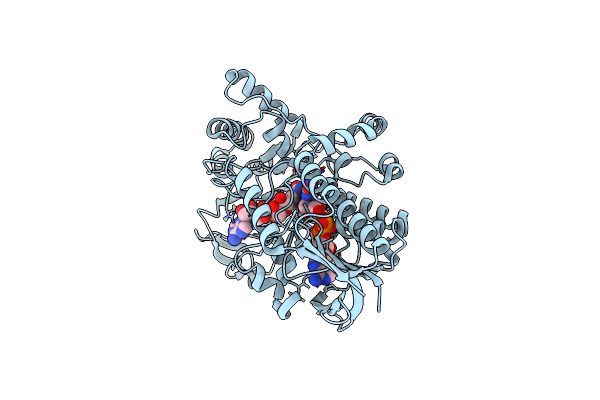 |
Structure Of Cyclohexanone Monooxygenase Mutant From Acinetobacter Calcoaceticus
Organism: Acinetobacter calcoaceticus
Method: X-RAY DIFFRACTION Resolution:2.63 Å Release Date: 2025-03-26 Classification: OXIDOREDUCTASE Ligands: FAD, NAD |
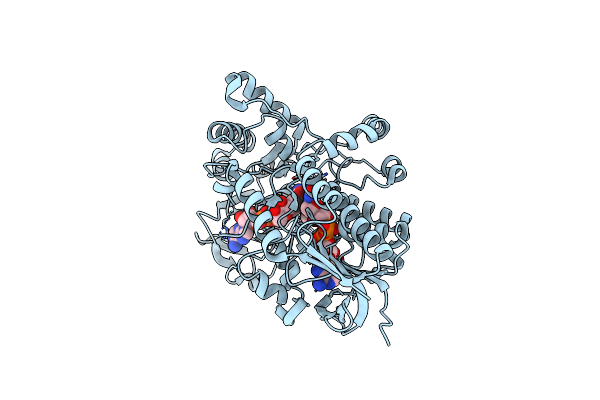 |
Structure Of Cyclohexanone Monooxygenase Mutant From Acinetobacter Calcoaceticus
Organism: Acinetobacter calcoaceticus
Method: X-RAY DIFFRACTION Resolution:2.02 Å Release Date: 2025-03-26 Classification: OXIDOREDUCTASE Ligands: NAI, FAD |
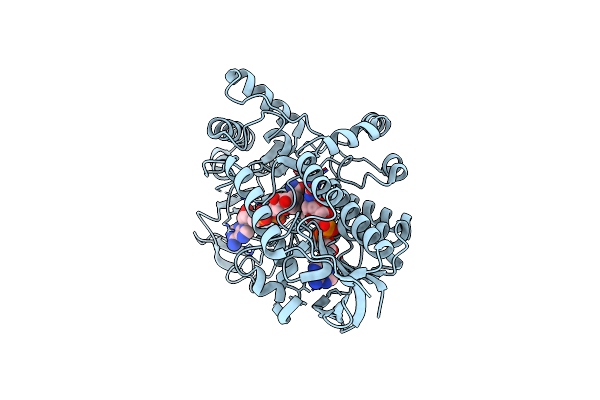 |
Structure Of Cyclohexanone Monooxygenase Mutant From Acinetobacter Calcoaceticus
Organism: Acinetobacter calcoaceticus
Method: X-RAY DIFFRACTION Resolution:2.52 Å Release Date: 2025-03-19 Classification: OXIDOREDUCTASE Ligands: FAD, NAI |
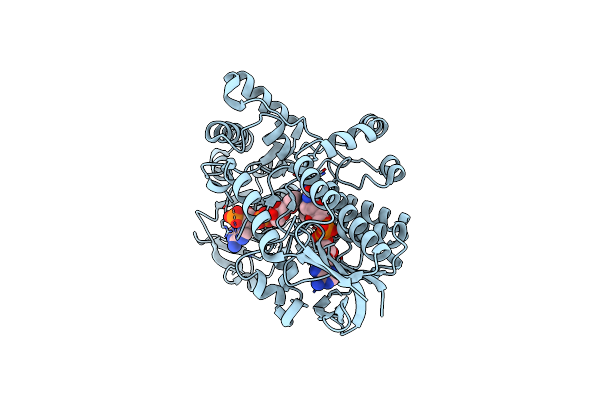 |
Structure Of Cyclohexanone Monooxygenase Mutant From Acinetobacter Calcoaceticus
Organism: Acinetobacter calcoaceticus
Method: X-RAY DIFFRACTION Resolution:1.67 Å Release Date: 2025-03-19 Classification: OXIDOREDUCTASE Ligands: FAD, NDP |
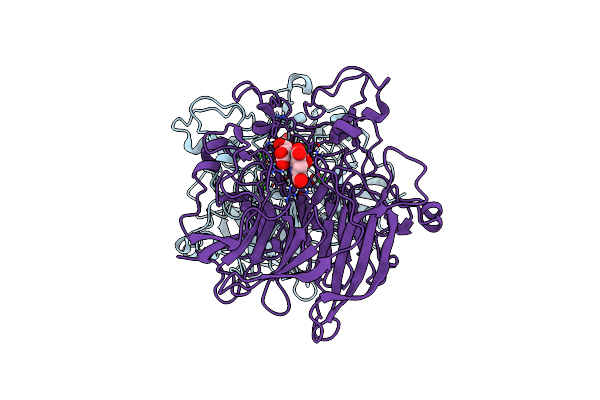 |
Soluble Glucose Dehydrogenase From Acinetobacter Calcoaceticus - Double Mutant Ph8
Organism: Acinetobacter calcoaceticus
Method: X-RAY DIFFRACTION Resolution:1.56 Å Release Date: 2024-05-08 Classification: OXIDOREDUCTASE Ligands: CA, A1H0D, LI |
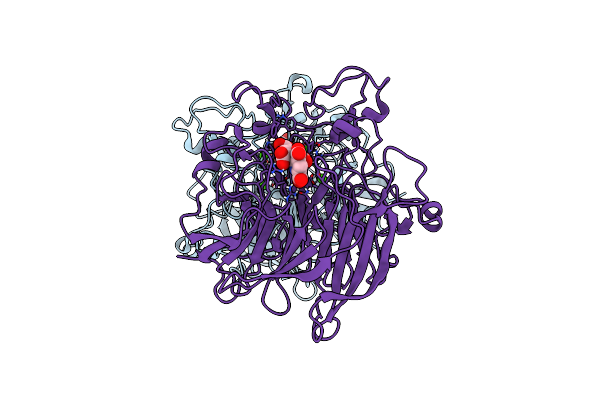 |
Soluble Glucose Dehydrogenase From Acinetobacter Calcoaceticus - Single Mutant Ph8
Organism: Acinetobacter calcoaceticus
Method: X-RAY DIFFRACTION Resolution:1.56 Å Release Date: 2024-05-08 Classification: OXIDOREDUCTASE Ligands: CA, A1H0D |
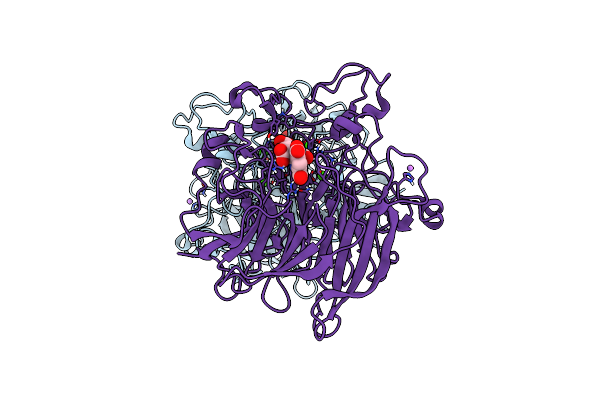 |
Soluble Glucose Dehydrogenase From Acinetobacter Calcoaceticus - Wild Type Ph8
Organism: Acinetobacter calcoaceticus
Method: X-RAY DIFFRACTION Resolution:1.19 Å Release Date: 2024-05-08 Classification: OXIDOREDUCTASE Ligands: A1H0D, CA, LI |
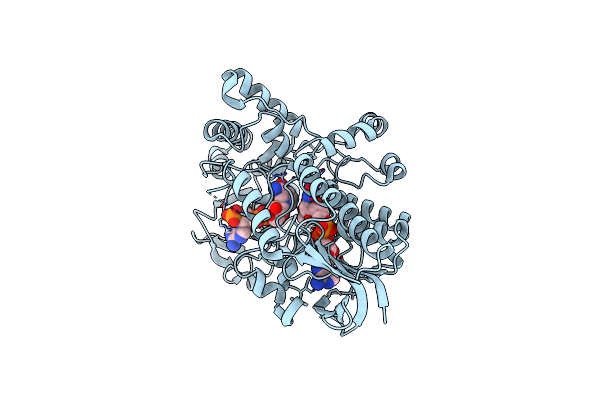 |
Structure Of Cyclohexanone Monooxygenase Mutant From Acinetobacter Calcoaceticus
Organism: Acinetobacter calcoaceticus
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2023-02-22 Classification: OXIDOREDUCTASE Ligands: FAD, NAP |
 |
Organism: Acinetobacter calcoaceticus
Method: X-RAY DIFFRACTION Resolution:2.42 Å Release Date: 2023-02-22 Classification: OXIDOREDUCTASE Ligands: FAD |
 |
Structure Of Cyclohexanone Monooxygenase Mutant From Acinetobacter Calcoaceticus
Organism: Acinetobacter calcoaceticus
Method: X-RAY DIFFRACTION Resolution:2.33 Å Release Date: 2023-02-15 Classification: OXIDOREDUCTASE Ligands: FAD, NAP |
 |
Organism: Acinetobacter calcoaceticus
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2022-09-07 Classification: DNA BINDING PROTEIN Ligands: IPH, ACT, ZN |
 |
X-Ray Structure Of Cyclohexanone Monooxygenase From Acinetobacter Calcoaceticus
Organism: Acinetobacter calcoaceticus
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2019-03-27 Classification: OXIDOREDUCTASE Ligands: FAD |
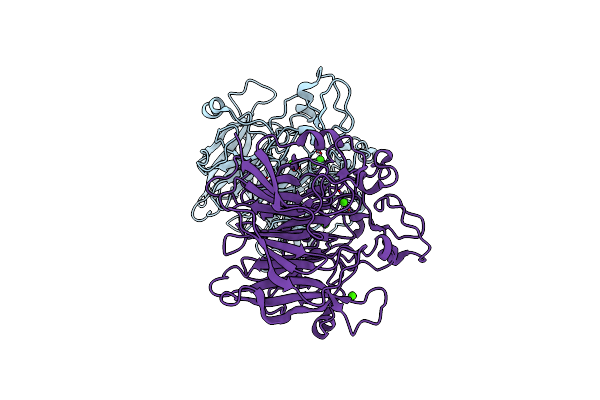 |
Apo Form Of The Soluble Pqq-Dependent Glucose Dehydrogenase From Acinetobacter Calcoaceticus
Organism: Acinetobacter calcoaceticus
Method: X-RAY DIFFRACTION Resolution:1.76 Å Release Date: 2017-12-20 Classification: OXIDOREDUCTASE Ligands: CA, CL |
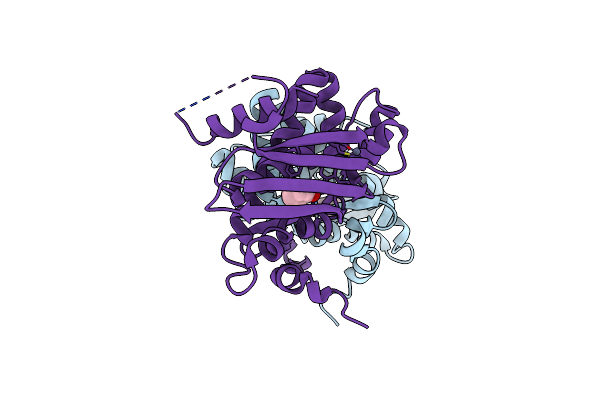 |
Crystal Structure Of The Aromatic Sensor Domain Of Mopr In Complex With Phenol
Organism: Acinetobacter calcoaceticus
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2016-07-13 Classification: TRANSCRIPTION Ligands: ZN, IPH |
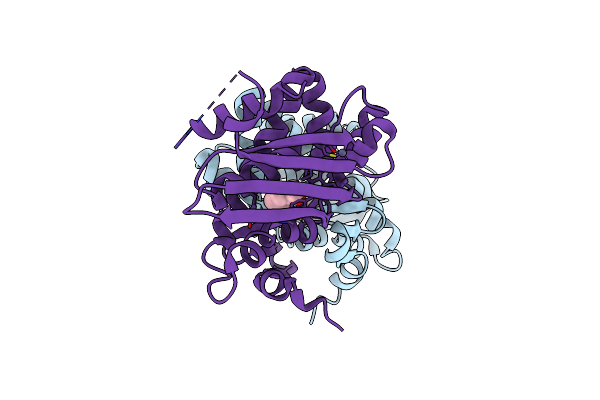 |
Crystal Structure Of The Aromatic Sensor Domain Of Mopr In Complex With Ocresol
Organism: Acinetobacter calcoaceticus
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2016-07-13 Classification: TRANSCRIPTION Ligands: ZN, JZ0 |
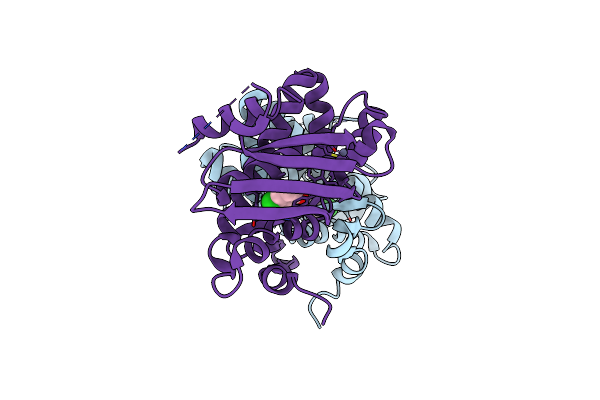 |
Crystal Structure Of The Aromatic Sensor Domain Of Mopr In Complex With 3-Chloro-Phenol
Organism: Acinetobacter calcoaceticus
Method: X-RAY DIFFRACTION Resolution:2.55 Å Release Date: 2016-07-13 Classification: TRANSCRIPTION Ligands: ZN, 3CH |

