Search Count: 520
All
Selected
 |
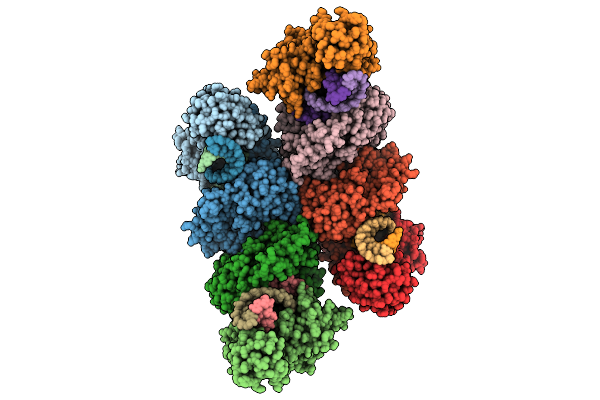
Cryo-Em Structure Of The Type Ii Secretion System Protein From Acidithiobacillus Caldus
Organism: Acidithiobacillus caldus (strain sm-1)
Method: ELECTRON MICROSCOPY Resolution:2.39 Å Release Date: 2025-11-19 Classification: TOXIN |
 |
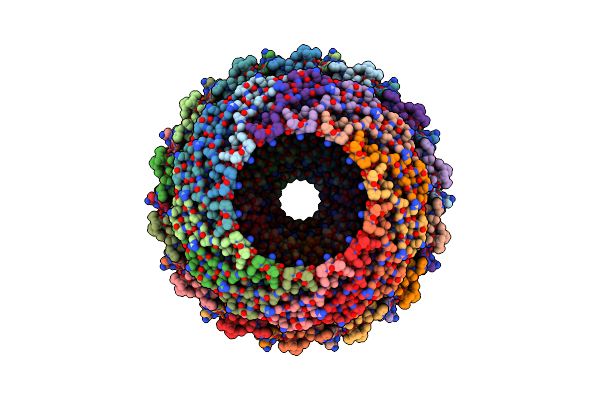
Cryo-Em Structure Of Filament Form Acidithiobacillus Caldus (Aca) Short Prokaryotic Argonautes, Hnh-Associated (Sparha) With Grna And Tdna
Organism: Acidithiobacillus caldus
Method: ELECTRON MICROSCOPY Release Date: 2025-10-22 Classification: IMMUNE SYSTEM/DNA/RNA Ligands: MN |
 |
Organism: Acidithiobacillus
Method: X-RAY DIFFRACTION Resolution:2.27 Å Release Date: 2025-05-14 Classification: LYASE Ligands: PLP |
 |
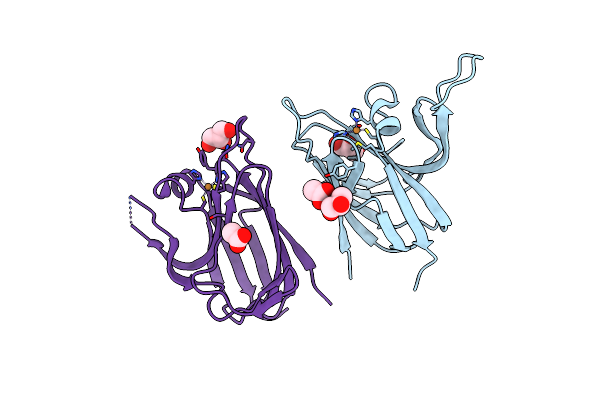
Cryo-Em Structure Of The Type Ii Secretion System Protein From Acidithiobacillus Caldus
Organism: Acidithiobacillus caldus (strain sm-1)
Method: ELECTRON MICROSCOPY Release Date: 2024-07-03 Classification: TOXIN |
 |
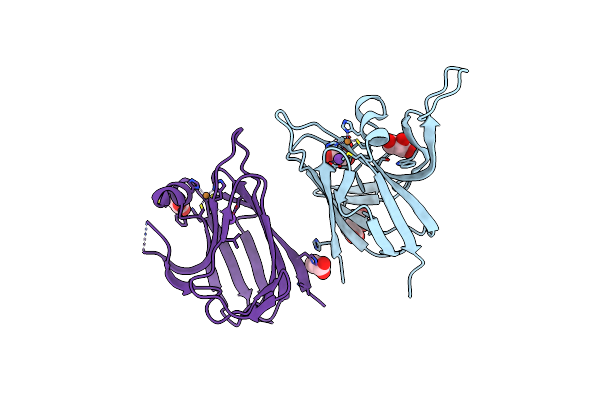
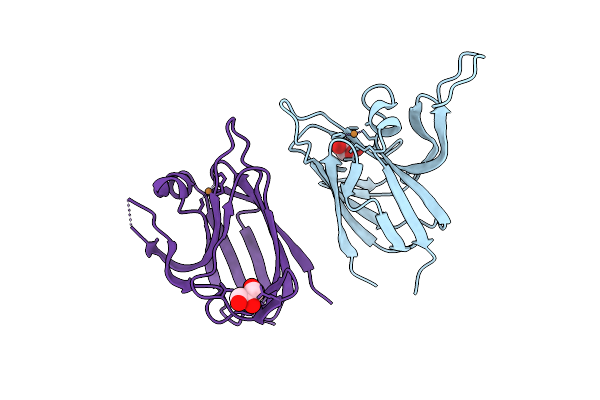
Crystal Structure Of The Cupredoxin Acop From Acidithiobacillus Ferrooxidans, Reduced Form
Organism: Acidithiobacillus ferrooxidans
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2023-09-13 Classification: METAL BINDING PROTEIN Ligands: CU1, GOL, ACT |
 |
Crystal Structure Of The Cupredoxin Acop From Acidithiobacillus Ferrooxidans, Oxidized Form
Organism: Acidithiobacillus ferrooxidans
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2023-09-13 Classification: METAL BINDING PROTEIN Ligands: CU, ACT, NA, CL, GOL |
 |
Crystal Structure Of The Cupredoxin Acop From Acidithiobacillus Ferrooxidans, H166A Mutant
Organism: Acidithiobacillus ferrooxidans
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2023-09-13 Classification: METAL BINDING PROTEIN Ligands: CU1, GOL |
 |
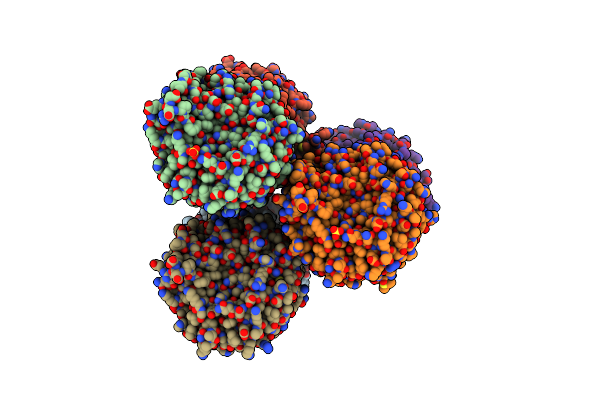
Crystal Structure Of The Cupredoxin Acop From Acidithiobacillus Ferrooxidans, M171A Mutant
Organism: Acidithiobacillus ferrooxidans
Method: X-RAY DIFFRACTION Resolution:1.82 Å Release Date: 2023-09-13 Classification: METAL BINDING PROTEIN Ligands: CU, ACT, GOL |
 |
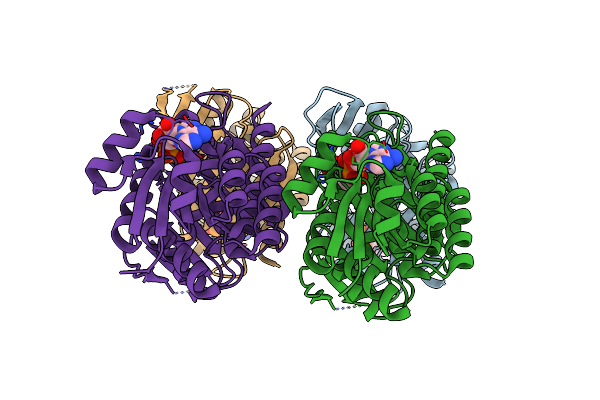
Organism: Acidithiobacillus ferrooxidans
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2021-01-27 Classification: HYDROLASE Ligands: SO4 |
 |
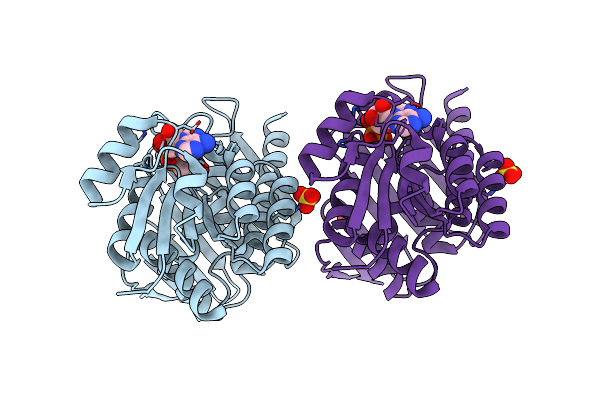
Udp-N-Acetylglucosamine 3-Dehydrogenase Gnna From Acidithiobacillus Ferrooxidans (P21)
Organism: Acidithiobacillus ferrooxidans (strain atcc 23270 / dsm 14882 / cip 104768 / ncimb 8455)
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2020-12-02 Classification: OXIDOREDUCTASE Ligands: NAD, F8U |
 |
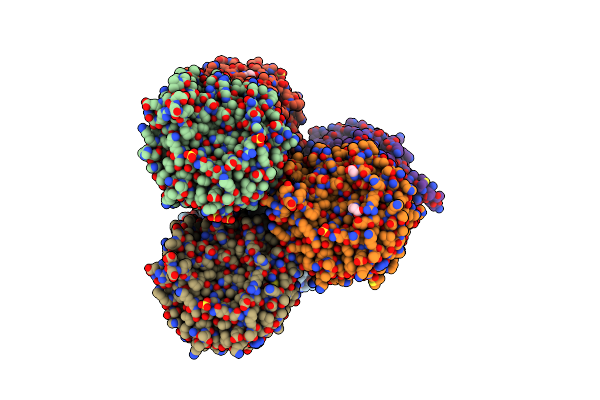
Udp-N-Acetylglucosamine 3-Dehydrogenase Gnna From Acidithiobacillus Ferrooxidans (P212121)
Organism: Acidithiobacillus ferrooxidans (strain atcc 23270 / dsm 14882 / cip 104768 / ncimb 8455)
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2020-12-02 Classification: OXIDOREDUCTASE Ligands: NAD, SO4 |
 |
Organism: Acidithiobacillus ferrooxidans
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2020-11-11 Classification: HYDROLASE Ligands: SO4, BAL, GLY |
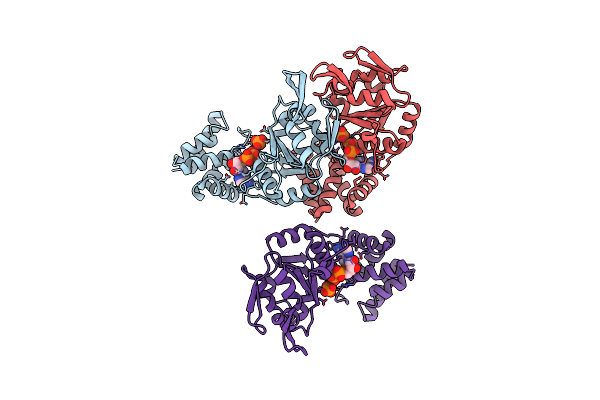 |
Crystal Structure Of Afcbbq2, A Moxr Aaa+-Atpase And Cbbqo-Type Rubisco Activase From Acidithiobacillus Ferrooxidans
Organism: Acidithiobacillus ferrooxidans atcc 23270
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2019-12-18 Classification: CHAPERONE Ligands: ADP, PO4 |
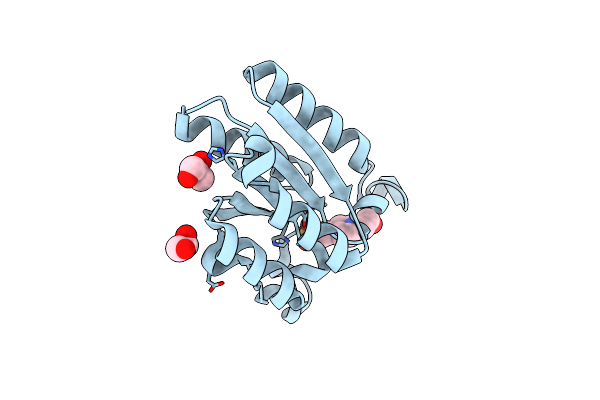 |
The Crystal Structure Of Type Ii Dehydroquinase From Acidithiobacillus Caldus Sm-1
Organism: Acidithiobacillus caldus (strain sm-1)
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2019-10-23 Classification: BIOSYNTHETIC PROTEIN Ligands: GOL, EPE |
 |
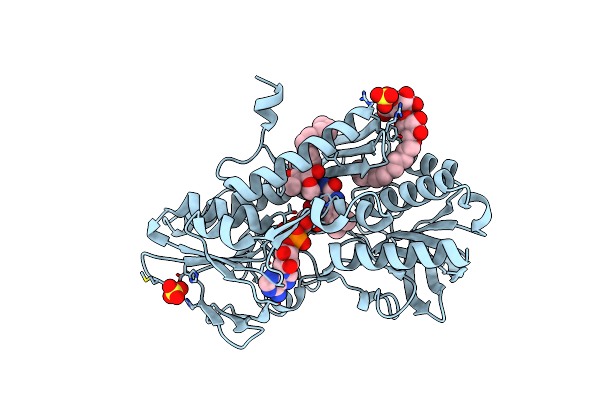
Structures Of Two Arsr As(Iii)-Responsive Repressors: Implications For The Mechanism Of Derepression
Organism: Acidithiobacillus ferrooxidans
Method: X-RAY DIFFRACTION Resolution:1.86 Å Release Date: 2019-07-03 Classification: TRANSCRIPTION Ligands: ARS, NA |
 |
Crystal Structure Of Sulfide:Quinone Oxidoreductase Cys356Ala Variant From Acidithiobacillus Ferrooxidans Complexed With Decylubiquinone
Organism: Acidithiobacillus ferrooxidans atcc 23270
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2012-05-16 Classification: OXIDOREDUCTASE Ligands: FAD, LMT, DCQ, SO4, H2S |
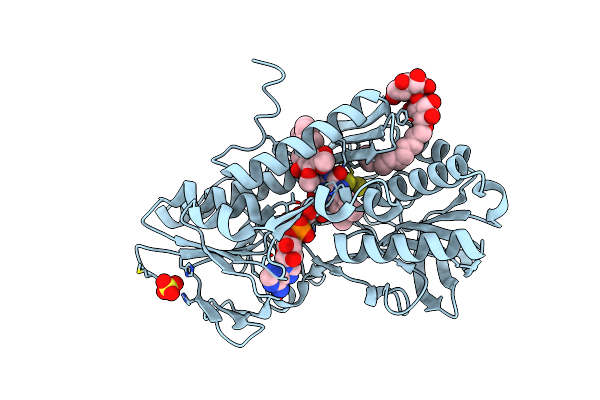 |
Crystal Structure Of Sulfide:Quinone Oxidoreductase Cys128Ala Variant From Acidithiobacillus Ferrooxidans Complexed With Decylubiquinone
Organism: Acidithiobacillus ferrooxidans
Method: X-RAY DIFFRACTION Resolution:2.18 Å Release Date: 2012-05-16 Classification: OXIDOREDUCTASE Ligands: FAD, LMT, DCQ, SO4, H2S |
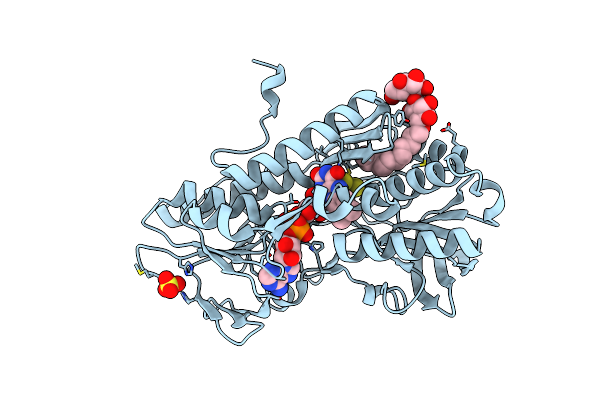 |
Crystal Structure Of Sulfide:Quinone Oxidoreductase Ser126Ala Variant From Acidithiobacillus Ferrooxidans
Organism: Acidithiobacillus ferrooxidans
Method: X-RAY DIFFRACTION Resolution:1.91 Å Release Date: 2012-05-16 Classification: OXIDOREDUCTASE Ligands: FAD, LMT, H2S, SO4 |
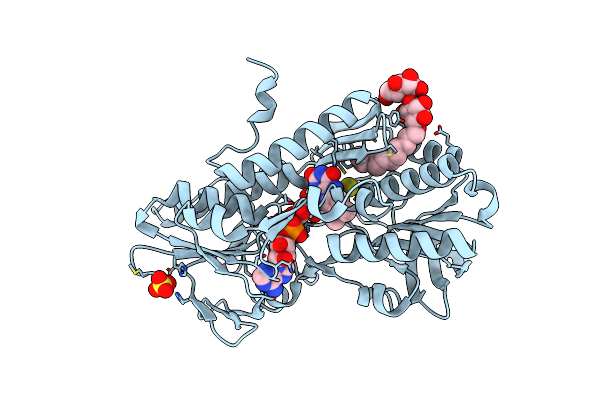 |
Crystal Structure Of Sulfide:Quinone Oxidoreductase Ser126Ala Variant From Acidithiobacillus Ferrooxidans Using 7.0 Kev Diffraction Data
Organism: Acidithiobacillus ferrooxidans
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2012-05-16 Classification: OXIDOREDUCTASE Ligands: FAD, LMT, H2S, SO4 |
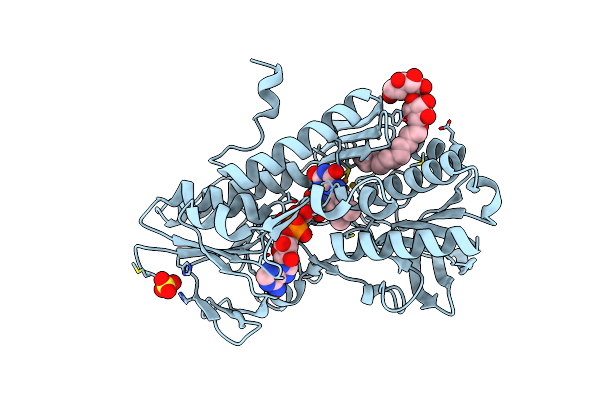 |
Crystal Structure Of Sulfide:Quinone Oxidoreductase From Acidithiobacillus Ferrooxidans In Complex With Sodium Selenide
Organism: Acidithiobacillus ferrooxidans
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2012-05-16 Classification: OXIDOREDUCTASE Ligands: FAD, LMT, SO4, 6SE, SE |

