Search Count: 55
 |
N-Acetylglucosamine 6-Phosphate Dehydratase: Inhibited 6-Phosphogluconic Acid State Of Nags
Organism: Streptomyces coelicolor
Method: X-RAY DIFFRACTION Resolution:1.69 Å Release Date: 2025-02-19 Classification: SUGAR BINDING PROTEIN Ligands: SO4, 6PG |
 |
Organism: Streptomyces coelicolor
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2025-02-19 Classification: SUGAR BINDING PROTEIN Ligands: SO4 |
 |
N-Acetylglucosamine 6-Phosphate Dehydratase: Glcnac6P Substrate-Bound State Of Nags
Organism: Streptomyces coelicolor
Method: X-RAY DIFFRACTION Resolution:2.59 Å Release Date: 2025-02-19 Classification: SUGAR BINDING PROTEIN Ligands: SO4, 16G |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2021-12-22 Classification: MOTOR PROTEIN Ligands: ADP |
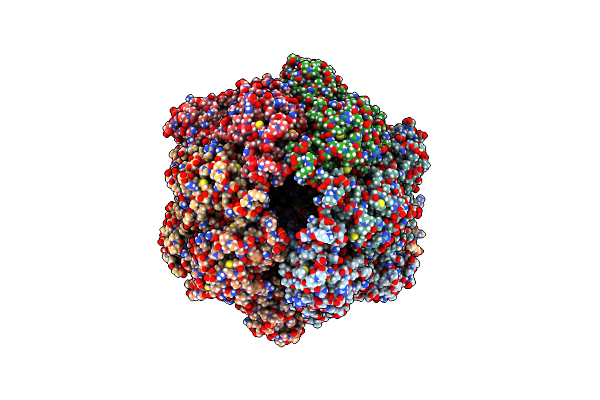 |
P2C-State Of Wild Type Human Mitochondrial Lonp1 Protease With Bound Endogenous Substrate Protein And In Presence Of Atp/Adp Mix
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2021-04-28 Classification: MOTOR PROTEIN Ligands: ATP, MG, ADP |
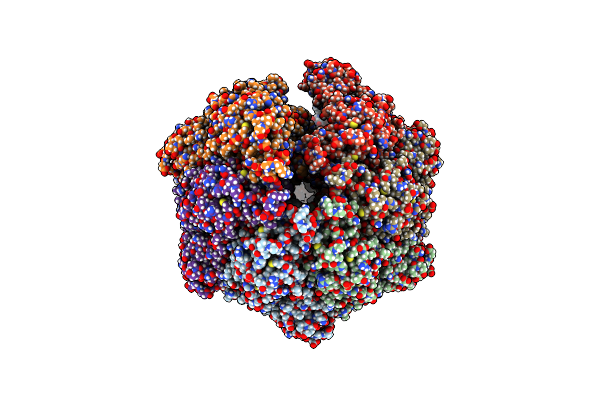 |
R-State Of Wild Type Human Mitochondrial Lonp1 Protease Bound To Endogenous Adp
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2021-04-28 Classification: MOTOR PROTEIN Ligands: ADP |
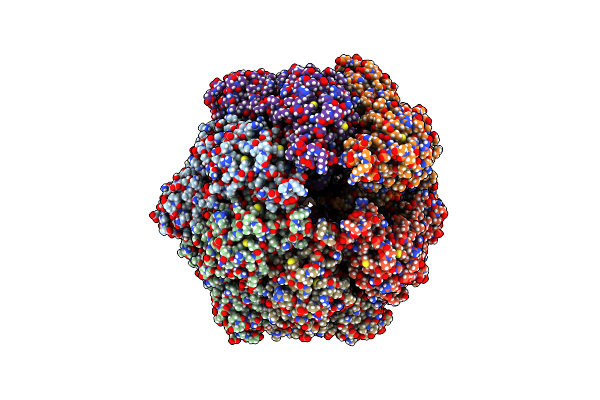 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2021-04-28 Classification: MOTOR PROTEIN Ligands: ADP |
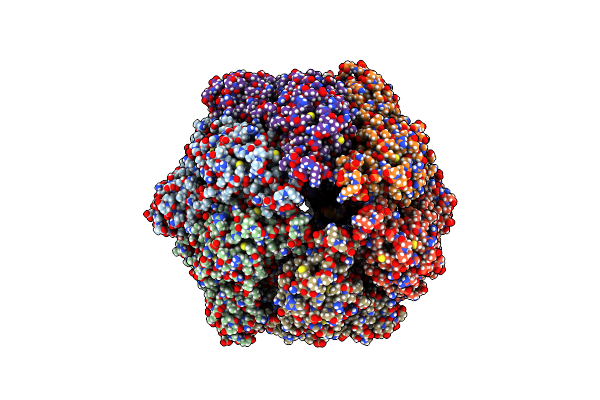 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2021-04-28 Classification: MOTOR PROTEIN Ligands: ADP |
 |
Organism: Staphylococcus aureus, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2021-04-14 Classification: TOXIN |
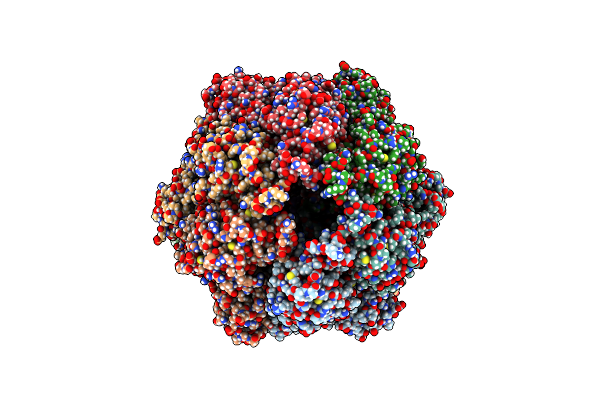 |
P2A-State Of Wild Type Human Mitochondrial Lonp1 Protease With Bound Substrate Protein And In Presence Of Atpgs
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2021-04-07 Classification: MOTOR PROTEIN Ligands: AGS, MG, ADP |
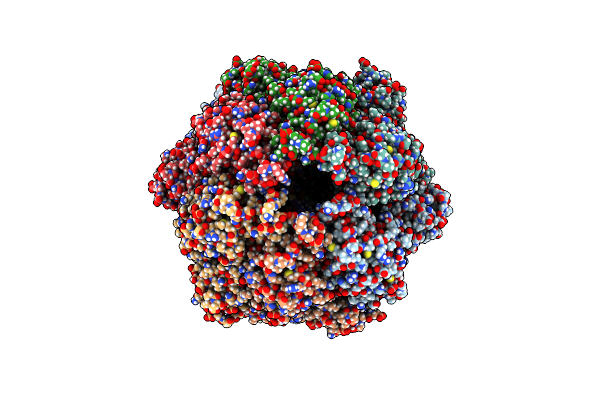 |
P1A-State Of Wild Type Human Mitochondrial Lonp1 Protease With Bound Substrate Protein And Atpgs
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2021-02-24 Classification: MOTOR PROTEIN Ligands: AGS, MG, ADP |
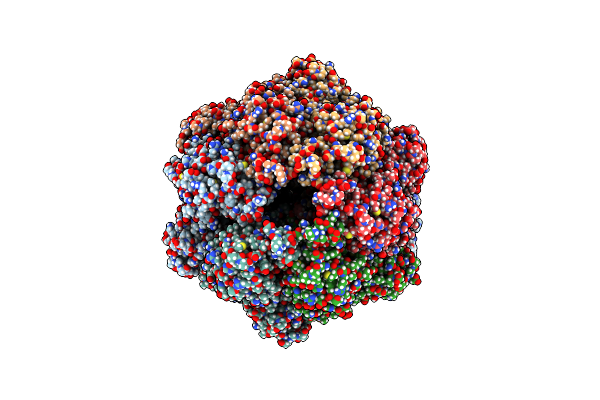 |
P1B-State Of Wild Type Human Mitochondrial Lonp1 Protease With Bound Endogenous Substrate Protein And In Presence Of Atp/Adp Mix
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2021-02-24 Classification: MOTOR PROTEIN Ligands: ATP, MG, ADP |
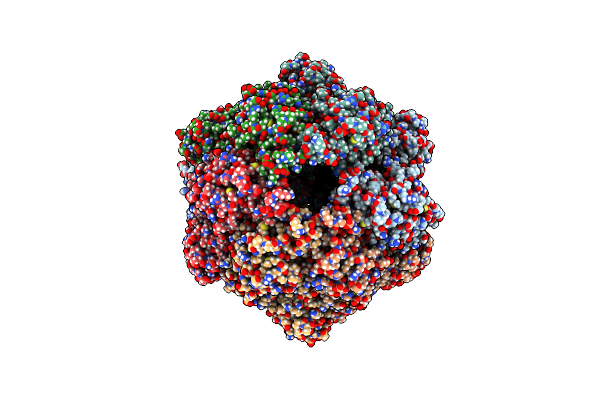 |
P1C-State Of Wild Type Human Mitochondrial Lonp1 Protease With Bound Substrate Protein In Presence Of Atp/Adp Mix
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2021-02-24 Classification: MOTOR PROTEIN Ligands: ATP, MG, ADP |
 |
Organism: Bos taurus
Method: ELECTRON CRYSTALLOGRAPHY Resolution:3.25 Å Release Date: 2021-01-27 Classification: HORMONE Ligands: ZN |
 |
3D Electron Diffraction Structure Of Thermolysin From Bacillus Thermoproteolyticus
Organism: Bacillus thermoproteolyticus
Method: ELECTRON CRYSTALLOGRAPHY Resolution:3.26 Å Release Date: 2021-01-27 Classification: HYDROLASE Ligands: ZN, CA |
 |
3D Electron Diffraction Structure Of Thaumatin From Thaumatococcus Daniellii
Organism: Thaumatococcus daniellii
Method: ELECTRON CRYSTALLOGRAPHY Resolution:2.76 Å Release Date: 2021-01-27 Classification: PLANT PROTEIN Ligands: CL |
 |
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2021-01-20 Classification: HORMONE Ligands: ZN, CL |
 |
Promiscuous Reductase Lugoii Catalyzes Keto-Reduction At C1 During Lugdunomycin Biosynthesis
Organism: Streptomyces sp. ql37
Method: X-RAY DIFFRACTION Resolution:1.08 Å Release Date: 2020-09-16 Classification: ANTIBIOTIC Ligands: NDP, EDO, PEG |
 |
Promiscuous Reductase Lugoii Catalyzes Keto-Reduction At C1 During Lugdunomycin Biosynthesis
Organism: Streptomyces sp. ql37
Method: X-RAY DIFFRACTION Resolution:1.08 Å Release Date: 2020-09-16 Classification: ANTIBIOTIC Ligands: EDO, NAP, P7K |
 |
Promiscuous Reductase Lugoii Catalyzes Keto-Reduction At C1 During Lugdunomycin Biosynthesis
Organism: Streptomyces sp. ql37
Method: X-RAY DIFFRACTION Resolution:1.57 Å Release Date: 2020-09-16 Classification: ANTIBIOTIC Ligands: NAP, P7Q, EDO |

