Search Count: 8
 |
Crystal Structure Of Maltose Binding Protein (Apo), Mutant Trp10 To 4-Cyanotryptophan
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.48 Å Release Date: 2024-12-18 Classification: SUGAR BINDING PROTEIN |
 |
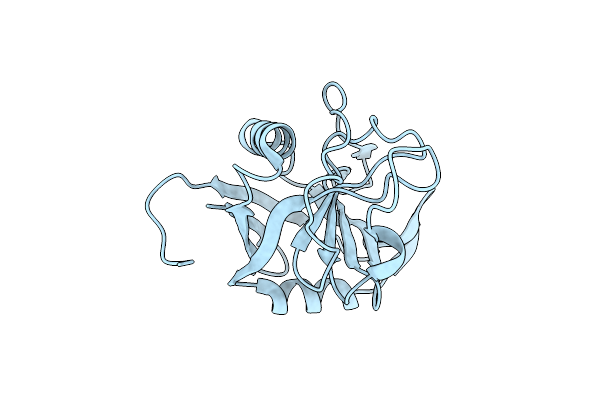
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.58 Å Release Date: 2024-12-18 Classification: SUGAR BINDING PROTEIN Ligands: PEG, EDO, PGE, NA, CD |
 |
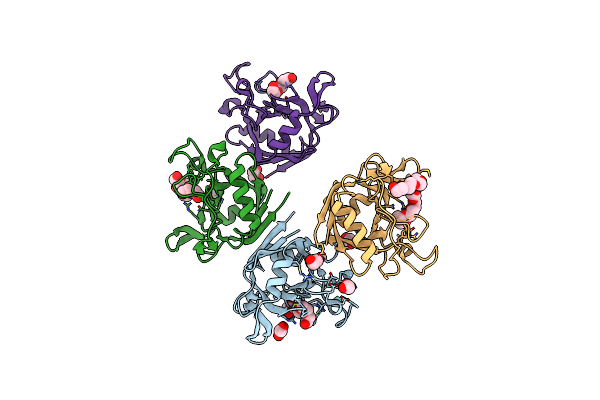
E. Coli Peptidyl-Prolyl Cis-Trans Isomerase Containing Delta1-Monofluoro-Leucines
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.22 Å Release Date: 2024-05-22 Classification: ISOMERASE |
 |
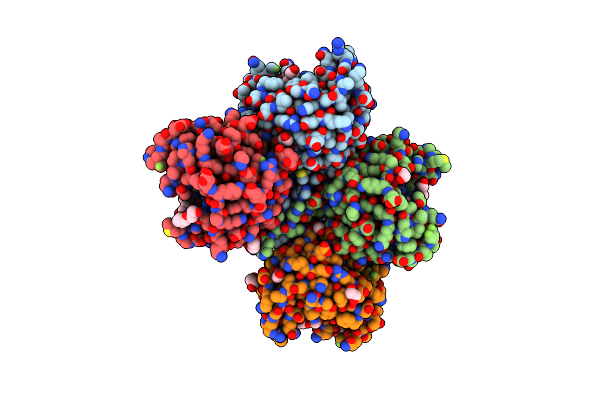
E. Coli Peptidyl-Prolyl Cis-Trans Isomerase Containing Delta2-Monofluoro-Leucines
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2024-05-22 Classification: ISOMERASE Ligands: EDO, PGE, PEG, 1PE, XPE |
 |
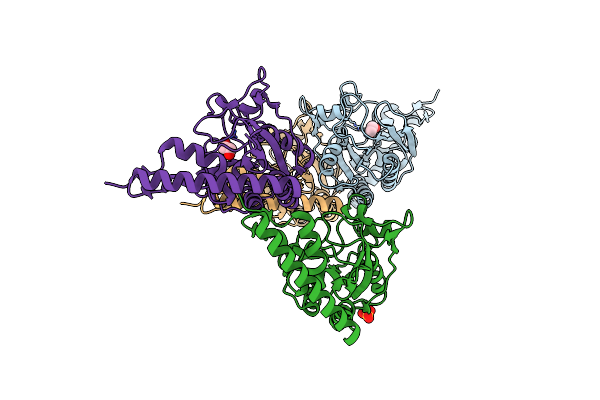
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2024-05-22 Classification: ISOMERASE Ligands: PEG, PGE, EDO, PG4, 2PE |
 |
Organism: Methanogenic archaeon mixed culture iso4-g1
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2022-03-02 Classification: TRANSLATION Ligands: EDO, SO4 |
 |
E. Coli Peptidyl-Prolyl Cis-Trans Isomerase, Mutant Phe27Cf3-Tyr/Phe98Cf3-Tyr
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2021-09-29 Classification: ISOMERASE |
 |
E. Coli Peptidyl-Prolyl Cis-Trans Isomerase, Mutant Phe4Ala Phe27Cf3-Phe/Phe98Cf3-Phe
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2021-09-29 Classification: ISOMERASE |

