Search Count: 15
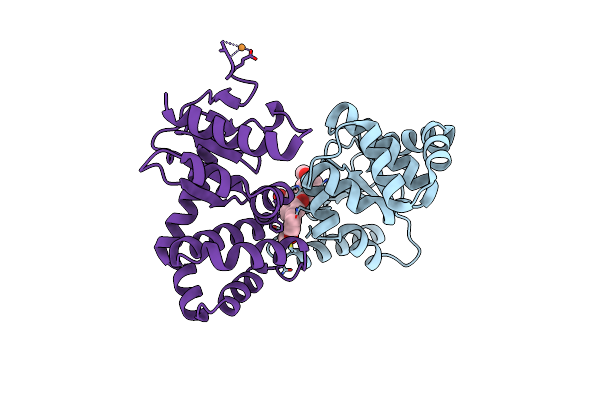 |
Crystal Structure Of E.Coli Dsba In Complex With 2-(2,6-Bis(3-Methoxyphenyl)Benzofuran-3-Yl)Acetic Acid
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2021-08-11 Classification: OXIDOREDUCTASE/INHIBITOR Ligands: VCY, CU |
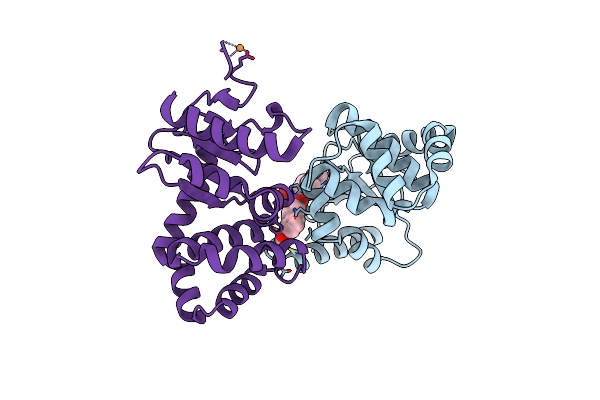 |
Crystal Structure Of E.Coli Dsba In Complex With 2-(6-(3-Methoxyphenyl)-2-(4-Methoxyphenyl)Benzofuran-3-Yl)Acetic Acid
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2021-08-11 Classification: OXIDOREDUCTASE/INHIBITOR Ligands: VE7, CU |
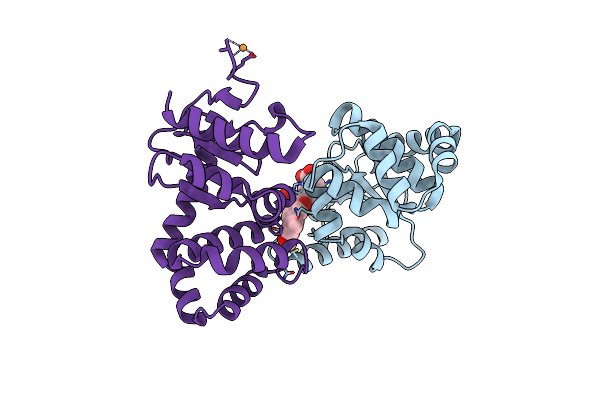 |
Crystal Structure Of E.Coli Dsba In Complex With 3-(3-(Carboxymethyl)-6-(3-Methoxyphenyl)Benzofuran-2-Yl)Benzoic Acid
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:1.99 Å Release Date: 2021-08-11 Classification: OXIDOREDUCTASE/INHIBITOR Ligands: VED, CU |
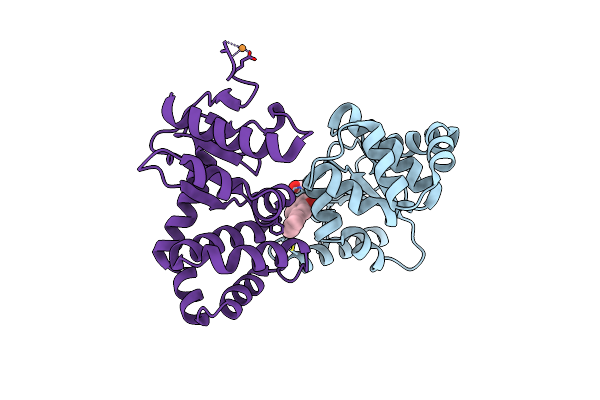 |
Crystal Structure Of Ecdsba In A Complex With 2-(6-Phenylbenzofuran-3-Yl)Acetic Acid
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:1.83 Å Release Date: 2021-08-11 Classification: OXIDOREDUCTASE/INHIBITOR Ligands: XPV, CU |
 |
Crystal Structure Of Ecdsba In A Complex With 2-(6-(3-Methoxyphenyl)Benzofuran-3-Yl)Acetic Acid
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2021-08-11 Classification: OXIDOREDUCTASE/INHIBITOR Ligands: XQ1, CU |
 |
Crystal Structure Of Ecdsba In A Complex With Methyl 2-(6-Bromo-2-Phenylbenzofuran-3-Yl)Acetate
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2021-08-11 Classification: OXIDOREDUCTASE/INHIBITOR Ligands: Y1G, CU |
 |
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2021-08-04 Classification: LYASE/LYASE INHIBITOR Ligands: YXP, CL |
 |
E.Coli Dsba In Complex With Benzofuran Compound 26 ([6-(3-Methoxyphenoxy)-1-Benzofuran-3-Yl]Acetic Acid)
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:1.74 Å Release Date: 2019-12-25 Classification: OXIDOREDUCTASE/OXIDOREDUCTASE INHIBITOR Ligands: OZG, DMS, PEG, CU |
 |
E.Coli Dsba In Complex With Benzofuran Compound 28 ((6-Benzyl-1-Benzofuran-3-Yl)Acetic Acid)
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:1.99 Å Release Date: 2019-12-25 Classification: OXIDOREDUCTASE/OXIDOREDUCTASE INHIBITOR Ligands: OZM, CU |
 |
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2019-11-06 Classification: OXIDOREDUCTASE/INHIBITOR Ligands: LD9, CU |
 |
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2019-11-06 Classification: OXIDOREDUCTASE/INHIBITOR Ligands: PEG, OR4, CU |
 |
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:1.67 Å Release Date: 2019-11-06 Classification: OXIDOREDUCTASE/INHIBITOR Ligands: OVG, CU |
 |
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:1.77 Å Release Date: 2019-11-06 Classification: OXIDOREDUCTASE/INHIBITOR Ligands: OVS, CU |
 |
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2019-11-06 Classification: OXIDOREDUCTASE/INHIBITOR Ligands: OVJ, CU |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.03 Å Release Date: 2015-01-21 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: 2SR, EDO, PEG, ZN, DMS, EPE |

