Search Count: 146
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-11-12 Classification: DNA Ligands: AU |
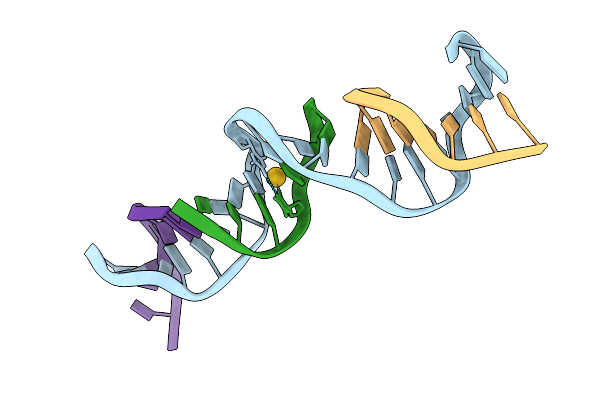 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-08-20 Classification: DNA Ligands: AU |
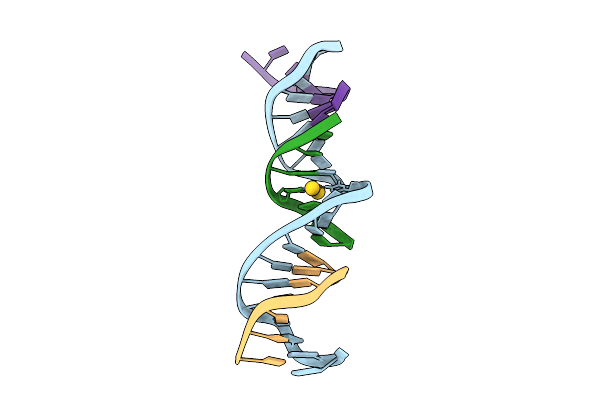 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-08-20 Classification: DNA Ligands: AU |
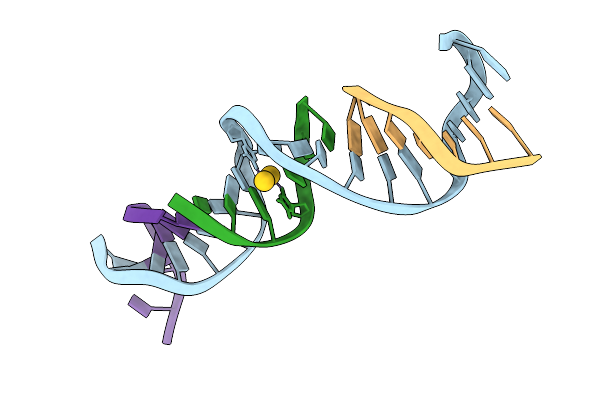 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-08-20 Classification: DNA Ligands: AU |
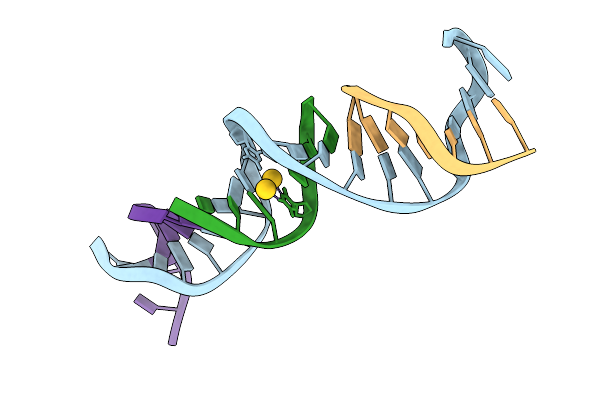 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-08-20 Classification: DNA Ligands: AU |
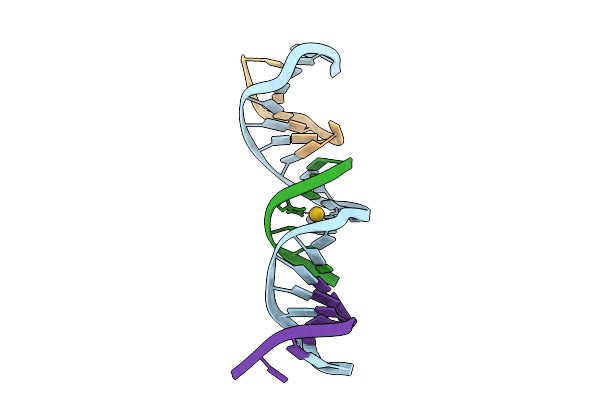 |
[F:Au+/Ag+:F-Ph8] Heterobimetallic Base Pair With Ag+ And Au+ Between A 2-Thio-Dt Homopair, Crystallized In The Presence Of Ag+, Au+ And Cu+
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-08-20 Classification: DNA Ligands: AG, AU |
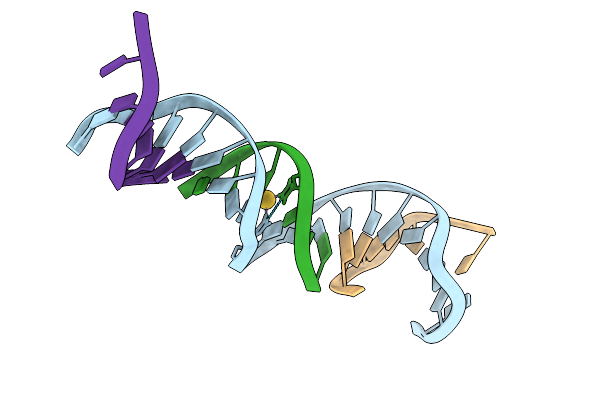 |
[Q:(Au3+):S_Ph11] Metal-Mediated Dna Base Pair In A Self-Assembling Rhombohedral Lattice
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-08-13 Classification: DNA Ligands: AU |
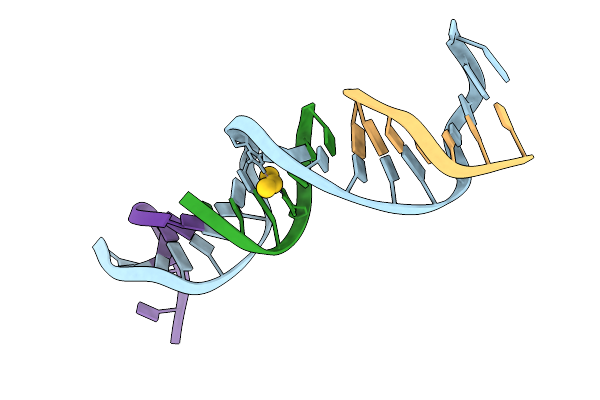 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-07-30 Classification: DNA Ligands: AU |
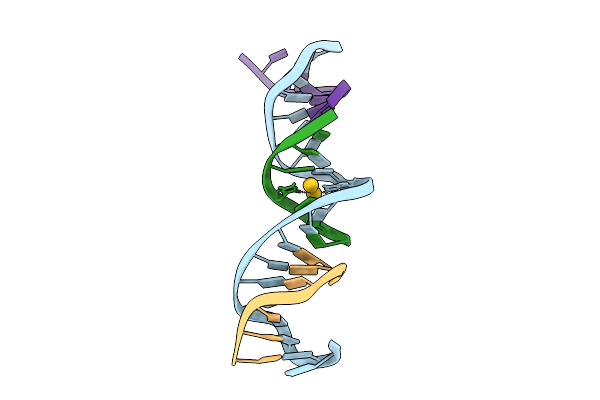 |
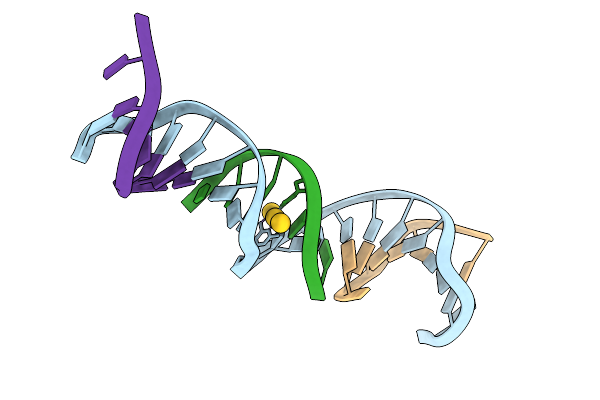
[D:Au3+:F] Tensegrity Triangle With A 2,6-Diaminopurine:2-Thiothymidine Base Pair Mediated By Au3+.
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-07-23 Classification: DNA Ligands: AU |
 |
[Q:(Au+)3:F_Ph11] Metal-Mediated Dna Base Pair In A Self-Assembling Rhombohedral Lattice
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-07-16 Classification: DNA Ligands: AU |
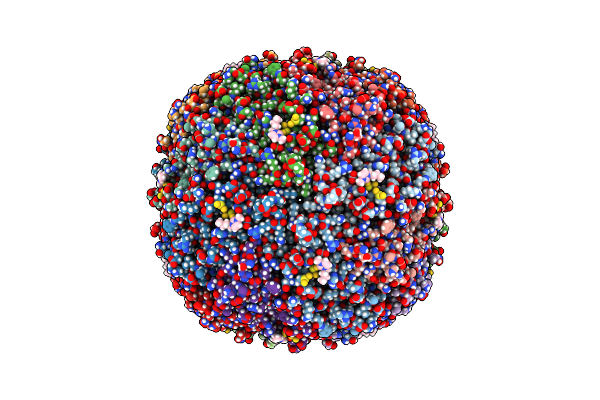 |
Structural Insights In The Huhf@Gold-Monocarbene Adduct: Aurophilicity Revealed In A Biological Context
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-05-07 Classification: METAL BINDING PROTEIN Ligands: BM0, AU |
 |
Crystal Structure Of The Adduct Formed Upon Reaction Of Aurothiomalate With Human Serum Transferrin (Apo-Form)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.02 Å Release Date: 2025-02-12 Classification: METAL TRANSPORT Ligands: CIT, AU, NAG |
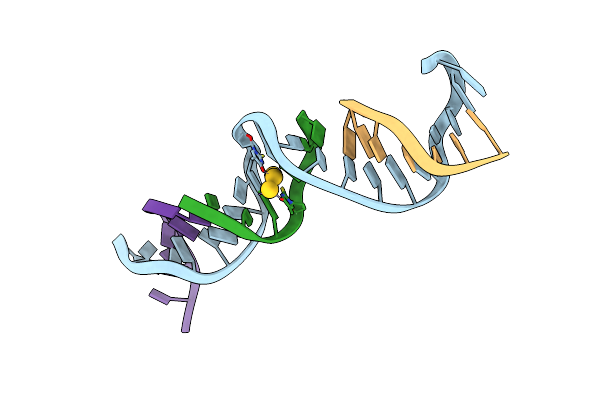 |
[F:3Au+:F] Metal-Mediated Dna Base Pair With 2-Thio-Thymidine In Tensegrity Triangle
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:5.86 Å Release Date: 2025-01-15 Classification: DNA Ligands: AU |
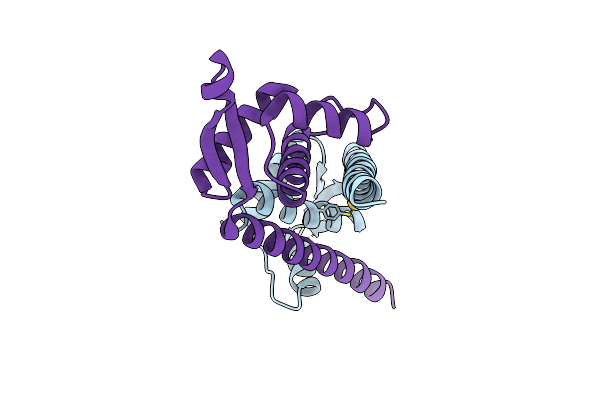 |
Crystal Structure Of Lmrr With V15 Replaced By Unnatural Amino Acid 4-Mercaptophenylalanine, Au(I) Bound
Organism: Lactococcus cremoris subsp. cremoris mg1363
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2024-12-25 Classification: METAL BINDING PROTEIN Ligands: AU |
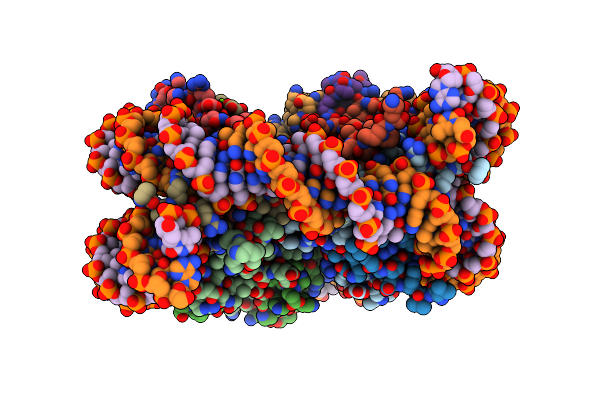 |
Structure Of Nucleosome Core With A Bound Metallopeptide Conjugate (Kaposi Sarcoma Associated Herpesvirus Lana Peptide-Au[I] Compound)
Organism: Homo sapiens, Human gammaherpesvirus 8
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2024-03-27 Classification: DNA Ligands: AU, XIS, MG |
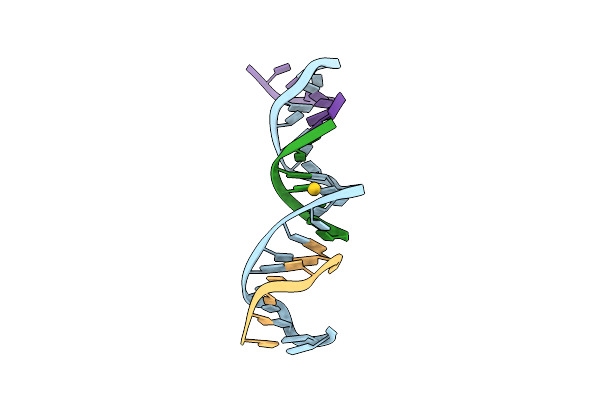 |
[T:Au+:T--(Ph11-Ph7; 5S)] Metal-Mediated Dna Base Pair In Tensegrity Triangle Grown At Ph 11 And Soaked In Ph 7 For 5S
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:4.59 Å Release Date: 2023-08-09 Classification: DNA Ligands: AU |
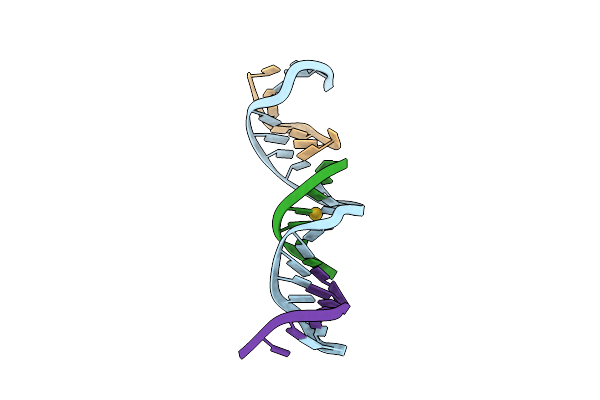 |
[T:Au+:T--(Ph11-Ph7; 15S)] Metal-Mediated Dna Base Pair In Tensegrity Triangle Grown At Ph 11 And Soaked In Ph 7 For 15S
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:4.52 Å Release Date: 2023-08-09 Classification: DNA Ligands: AU |
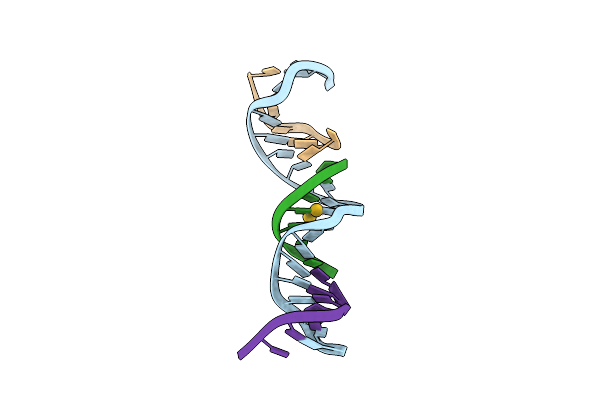 |
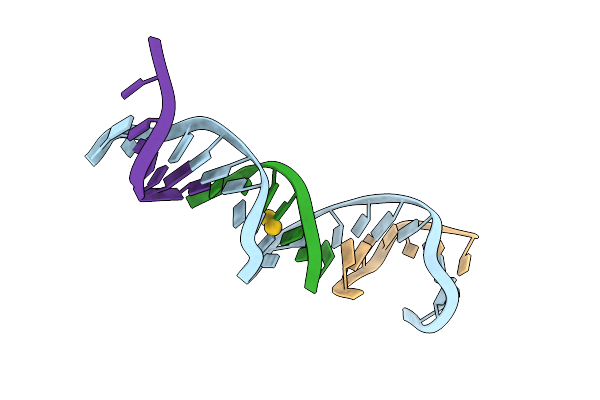
[T:Au+:T--(Ph11-Ph7; 45S)] Metal-Mediated Dna Base Pair In Tensegrity Triangle Grown At Ph 11 And Soaked In Ph 7 For 45S
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:4.07 Å Release Date: 2023-08-09 Classification: DNA Ligands: AU |
 |
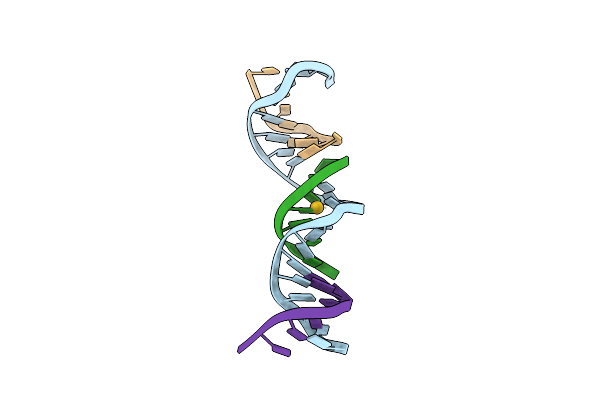
[T:Au+:T--(Ph11-Ph7; 180S)] Metal-Mediated Dna Base Pair In Tensegrity Triangle Grown At Ph 11 And Soaked In Ph 7 For 180S
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:4.87 Å Release Date: 2023-08-09 Classification: DNA Ligands: AU |
 |
[T:Au+:T--(Ph11-Ph7; 420S)] Metal-Mediated Dna Base Pair In Tensegrity Triangle Grown At Ph 11 And Soaked In Ph 7 For 420S
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:4.71 Å Release Date: 2023-08-09 Classification: DNA Ligands: AU |

