Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
 |
Structure Of Rv2985, A Nudix Diadenosine Polyphosphatase From M. Tuberculosis, In Complex With Putative Atp
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Release Date: 2026-01-14 Classification: CELL INVASION Ligands: ATP, SO4 |
 |
Hexamer Msp1 From S.Cerevisiae (With A Catalytic Dead Mutation) In Complex With An Unknown Peptide Substrate
Organism: Saccharomyces cerevisiae s288c, Escherichia coli bl21(de3)
Method: ELECTRON MICROSCOPY Release Date: 2026-01-14 Classification: MEMBRANE PROTEIN Ligands: ATP, MG |
 |
Organism: Penicillium aurantiogriseum
Method: X-RAY DIFFRACTION Release Date: 2026-01-14 Classification: BIOSYNTHETIC PROTEIN Ligands: GOL, TRS, MG, ATP |
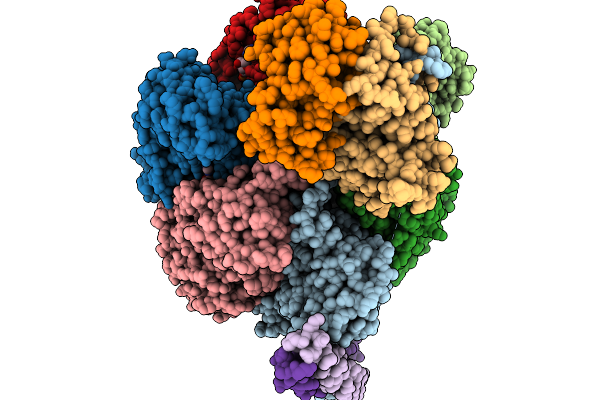 |
Cryo-Em Structure Of F-Atp Synthase From Mycobacteroides Abscessus (Rotational State 1)
Organism: Mycobacteroides abscessus subsp. abscessus
Method: ELECTRON MICROSCOPY Release Date: 2026-01-14 Classification: HYDROLASE Ligands: ATP, MG, ADP |
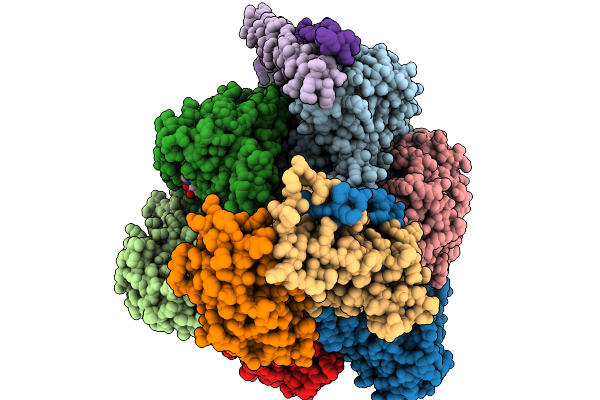 |
Cryo-Em Structure Of F-Atp Synthase From Mycobacteroides Abscessus (Rotational State 2)
Organism: Mycobacteroides abscessus subsp. abscessus
Method: ELECTRON MICROSCOPY Release Date: 2026-01-14 Classification: HYDROLASE Ligands: ATP, MG, ADP |
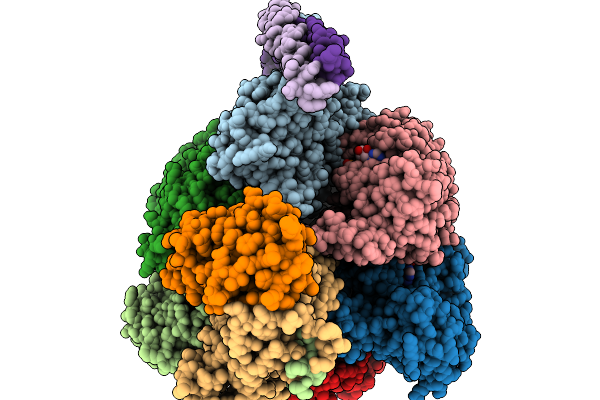 |
Cryo-Em Structure Of F-Atp Synthase From Mycobacteroides Abscessus (Rotational State 3)
Organism: Mycobacteroides abscessus subsp. abscessus
Method: ELECTRON MICROSCOPY Release Date: 2026-01-14 Classification: HYDROLASE Ligands: ATP, MG, ADP |
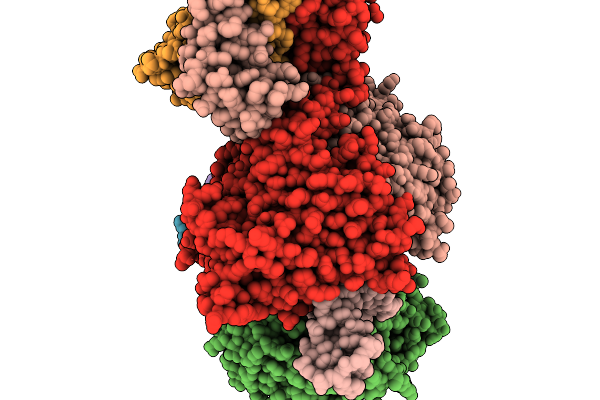 |
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2026-01-07 Classification: DNA BINDING PROTEIN/RNA/DNA Ligands: ATP, MG |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-31 Classification: HYDROLASE Ligands: ATP, ADP |
 |
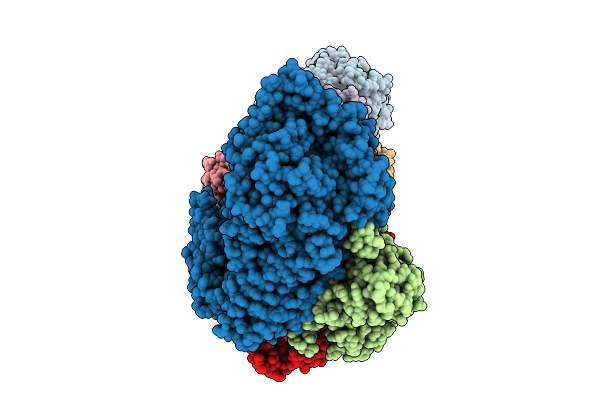
Hexamer Msp1 From S.Cerevisiae(With A Catalytic Dead Mutation) In Complex With An Unknown Peptide Substrate
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c), Escherichia coli bl21(de3)
Method: ELECTRON MICROSCOPY Release Date: 2025-12-31 Classification: MEMBRANE PROTEIN Ligands: ATP, MG |
 |
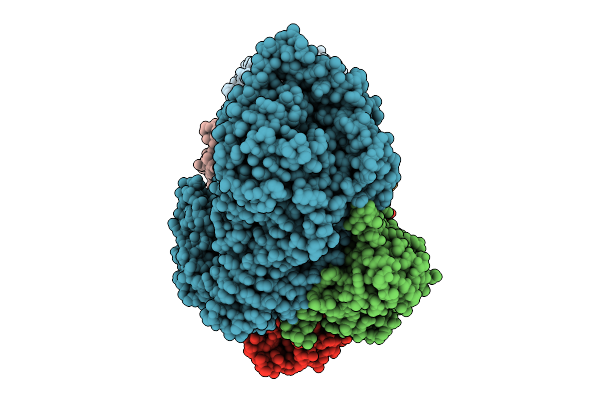
Organism: Archaeoglobus fulgidus dsm 4304, Pyrococcus furiosus dsm 3638
Method: ELECTRON MICROSCOPY Release Date: 2025-12-31 Classification: ANTIVIRAL PROTEIN Ligands: MG, ATP, ZN |
 |
Organism: Archaeoglobus fulgidus dsm 4304, Archaeoglobus fulgidus, Pyrococcus furiosus dsm 3638
Method: ELECTRON MICROSCOPY Release Date: 2025-12-31 Classification: ANTIVIRAL PROTEIN Ligands: MG, ATP, ZN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-31 Classification: HYDROLASE Ligands: ATP, ADP |
 |
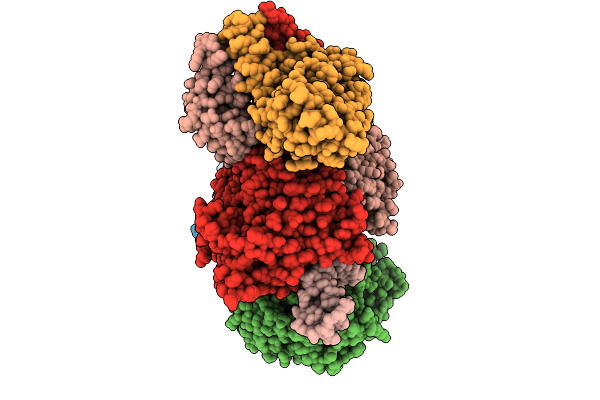
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-31 Classification: HYDROLASE Ligands: ATP, ADP |
 |
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2025-12-31 Classification: DNA BINDING PROTEIN/RNA/DNA Ligands: ATP, MG |
 |
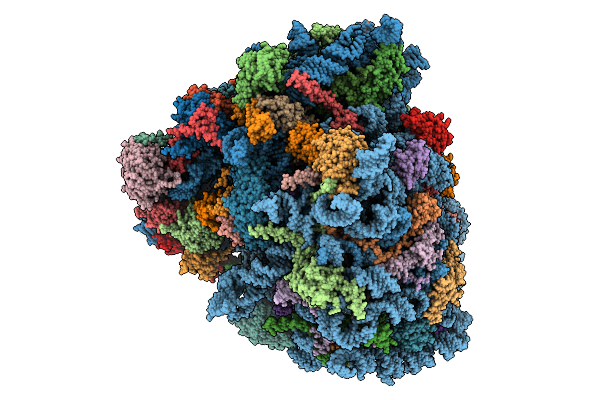
Gtpbp1*Gcp*Phe-Trna*Ribosome In The Gtpase Activation-Like State, Structure Iii
Organism: Homo sapiens, Oryctolagus cuniculus, Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2025-12-31 Classification: RIBOSOME Ligands: SPD, ZN, GTP, PHE, ATP, MET, K, GCP, MG |
 |
Gtpbp1*Gdp*Phe-Trna*Ribosome In The Post-Gtp Hydrolysis State, Structure Iv
Organism: Homo sapiens, Oryctolagus cuniculus, Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2025-12-31 Classification: RIBOSOME Ligands: SPD, ZN, GTP, PHE, ATP, MET, K, GDP, MG |
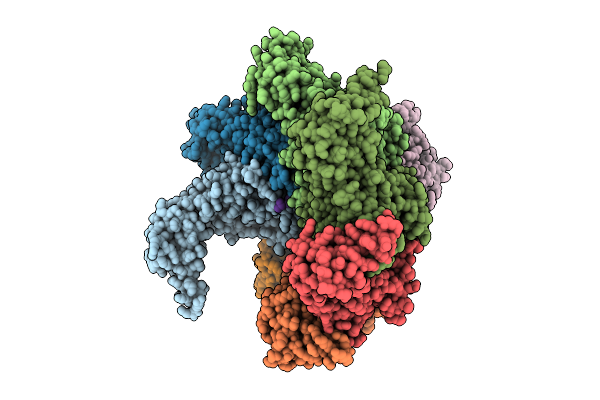 |
Octamer Msp1 From S.Cerevisiae(With A Catalytic Dead Mutation) In Complex With An Unknown Peptide Substrate
Organism: Saccharomyces cerevisiae s288c
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: MEMBRANE PROTEIN Ligands: MG, ATP |
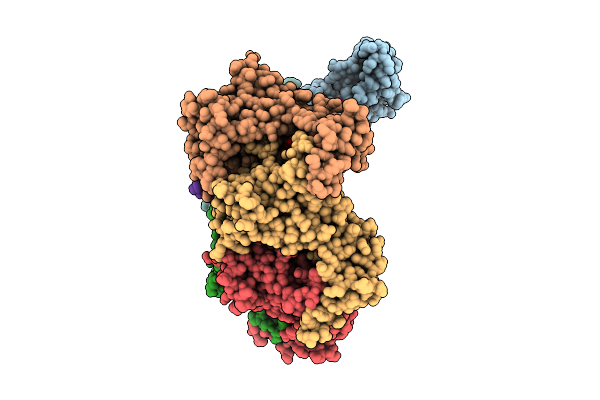 |
Hexamer Msp1 From S.Cerevisiae (With A Catalytic Dead Mutation) In Complex With An Unknown Peptide Substrate
Organism: Escherichia coli bl21(de3), Saccharomyces cerevisiae s288c
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: MEMBRANE PROTEIN Ligands: ATP, MG |
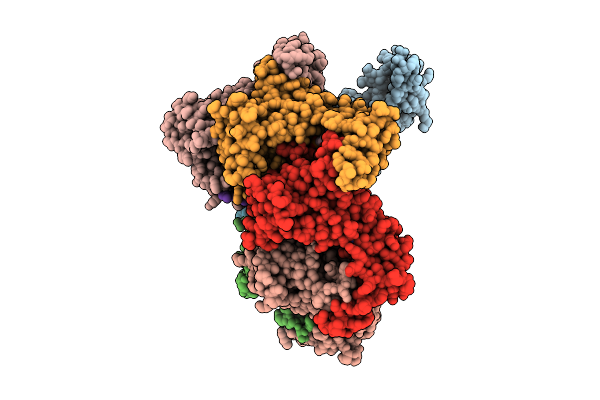 |
Heptamer Msp1 From S.Cerevisiae(With A Catalytic Dead Mutation) In Complex With An Unknown Peptide Substrate
Organism: Saccharomyces cerevisiae s288c
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: MEMBRANE PROTEIN Ligands: ATP, MG |
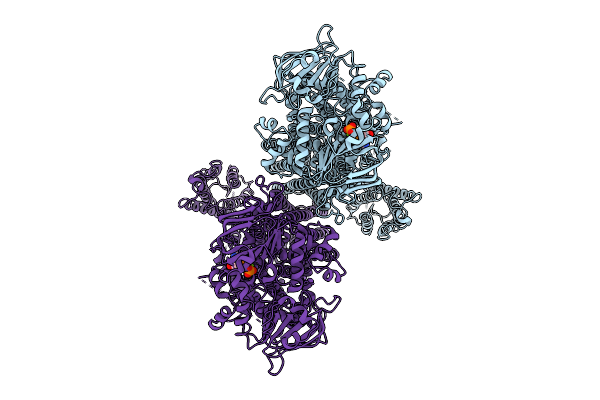 |
Organism: Arabidopsis thaliana
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: TRANSPORT PROTEIN Ligands: ATP, MG |