Search Count: 52
 |
Closed Conformation Of Arsa From L. Ferriphilum In Complex With Mgatp And Arsenite
Organism: Leptospirillum ferriphilum
Method: ELECTRON MICROSCOPY Release Date: 2025-08-20 Classification: HYDROLASE Ligands: ATP, MG, ARS |
 |
Closed Conformation Of Arsa From L. Ferriphilum In Complex With Mgatp And Arsenite At 1.5 Minute Time Point
Organism: Leptospirillum ferriphilum ml-04
Method: ELECTRON MICROSCOPY Release Date: 2025-08-20 Classification: HYDROLASE Ligands: ATP, MG, ARS |
 |
Crystal Structure Of Triosephosphate Isomerase From Leishmania Orientalis At 1.45 Angstroms Resolution With An Arsenic Atom Bound At Cys57
Organism: Leishmania orientalis
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2024-05-15 Classification: ISOMERASE Ligands: ARS |
 |
Crystal Structure Of Triosephosphate Isomerase From Leishmania Orientalis At 1.88A With An Arsenic Ion Bound At Cys57
Organism: Leishmania orientalis
Method: X-RAY DIFFRACTION Resolution:1.88 Å Release Date: 2024-05-01 Classification: ISOMERASE Ligands: ARS |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.02 Å Release Date: 2022-08-31 Classification: TRANSCRIPTION Ligands: ARS, ZN |
 |
Human P53 Core Domain With Germline Hot Spot Mutation M133T In Complex With The Natural Pig3 P53-Response Element And Arsenic
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.77 Å Release Date: 2022-06-22 Classification: TRANSCRIPTION Ligands: ARS, ZN |
 |
Human P53 Core Domain With Hot Spot Mutation R282W In Complex With The Natural Cdkn1A(P21) P53-Response Element And Arsenic
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2022-06-22 Classification: TRANSCRIPTION Ligands: ARS, ZN, PEG |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2021-03-03 Classification: TRANSCRIPTION Ligands: ARS, ZN, GOL, SO4 |
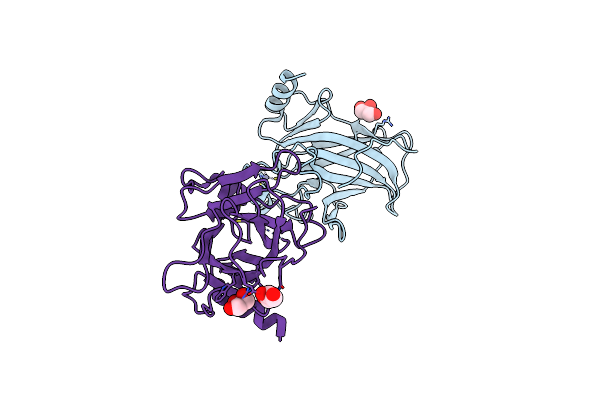 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.74 Å Release Date: 2021-03-03 Classification: TRANSCRIPTION Ligands: GOL, ARS, ZN |
 |
Crystal Structure Of A Self-Assembling Dna Scaffold Containing Ta Sticky Ends And Rhombohedral Symmetry
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2020-09-23 Classification: DNA Ligands: ARS |
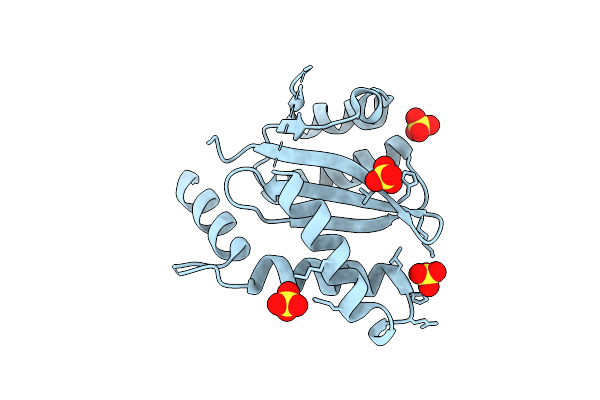 |
Crystal Structure Of Hiv-1 Integrase Catalytic Core Domain (A128T/K173Q/F185K)
Organism: Human immunodeficiency virus 1
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2020-07-01 Classification: VIRAL PROTEIN Ligands: SO4, ARS |
 |
Structures Of Two Arsr As(Iii)-Responsive Repressors: Implications For The Mechanism Of Derepression
Organism: Acidithiobacillus ferrooxidans
Method: X-RAY DIFFRACTION Resolution:1.86 Å Release Date: 2019-07-03 Classification: TRANSCRIPTION Ligands: ARS, NA |
 |
Structures Of Two Arsr As(Iii)-Responsive Repressors: Implications For The Mechanism Of Derepression
Organism: Corynebacterium glutamicum atcc 13032
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2019-07-03 Classification: TRANSCRIPTION Ligands: ARS, MPD |
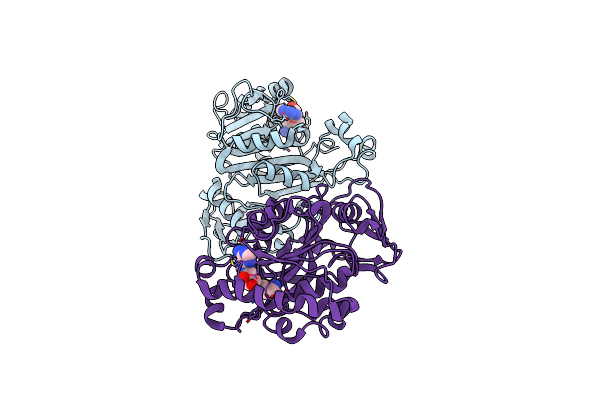 |
The Structure Of An As(Iii) S-Adenosylmethionine Methyltransferase With As(Iii) And S-Adenosyl-L-Homocysteine (Sah)
Organism: Cyanidioschyzon sp. 5508
Method: X-RAY DIFFRACTION Resolution:2.84 Å Release Date: 2018-06-27 Classification: TRANSFERASE Ligands: SAH, ARS |
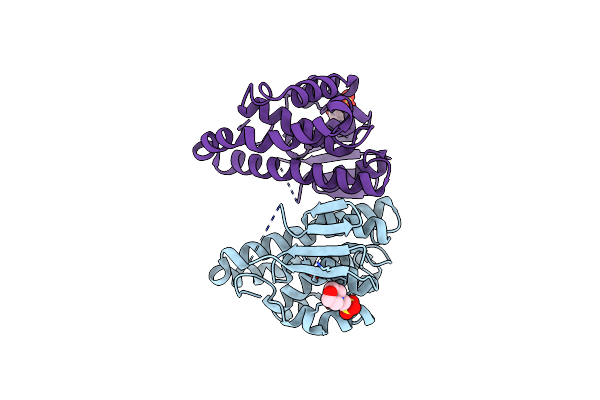 |
Crystal Structure Of Human Glutathione Transferase Pi Complexed With A Metalloid In The Absence Of Glutathione
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.11 Å Release Date: 2016-12-07 Classification: TRANSFERASE Ligands: MES, ARS |
 |
Organism: Pseudomonas aeruginosa (strain atcc 15692 / pao1 / 1c / prs 101 / lmg 12228)
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2015-09-23 Classification: OXIDOREDUCTASE Ligands: ARS, MG, HEM |
 |
[Fefe]-Hydrogenase Oxygen Inactivation Is Initiated By The Modification And Degradation Of The H Cluster 2Fe Subcluster
Organism: Chlamydomonas reinhardtii
Method: X-RAY DIFFRACTION Resolution:2.29 Å Release Date: 2015-01-28 Classification: OXIDOREDUCTASE Ligands: SF4, CL, ARS |
 |
Crystal Structure Analysis Of Leishmania Siamensis Triosephosphate Isomerase
Organism: Leishmania sp. siamensis
Method: X-RAY DIFFRACTION Resolution:1.93 Å Release Date: 2013-09-11 Classification: ISOMERASE Ligands: ARS, NA |
 |
Hiv-1 Integrase Catalytic Core Domain A128T Mutant Complexed With Allosteric Inhibitor
Organism: Human immunodeficiency virus type 1 (new york-5 isolate)
Method: X-RAY DIFFRACTION Resolution:2.16 Å Release Date: 2013-05-01 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: ARS, LF2 |
 |
Organism: Human immunodeficiency virus type 1 (new york-5 isolate)
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2013-05-01 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: ARS, LF2 |

