Search Count: 287
 |
Organism: Streptomyces monomycini
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: BIOSYNTHETIC PROTEIN Ligands: HEM, ARG |
 |
Organism: Streptomyces monomycini
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: BIOSYNTHETIC PROTEIN Ligands: HEM, ARG |
 |
Organism: Streptomyces monomycini
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: BIOSYNTHETIC PROTEIN Ligands: HEM, ARG |
 |
Organism: Streptomyces monomycini
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: BIOSYNTHETIC PROTEIN Ligands: ARG, HEM |
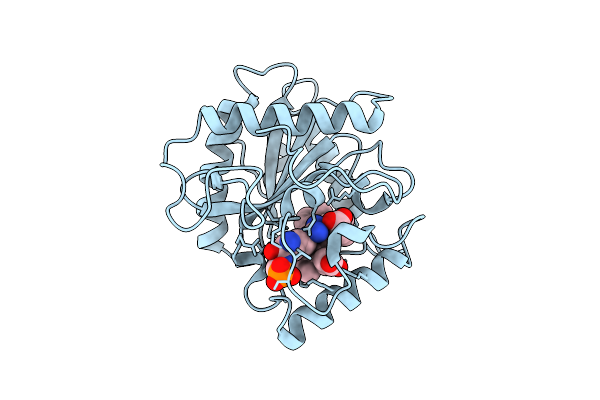 |
Organism: Streptomyces monomycini
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: BIOSYNTHETIC PROTEIN Ligands: HEM, ARG, PGL |
 |
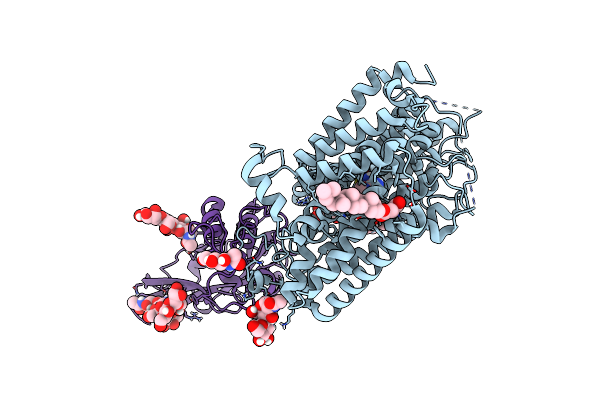
Organism: Pseudomonas aeruginosa
Method: ELECTRON MICROSCOPY Release Date: 2025-08-13 Classification: TRANSPORT PROTEIN Ligands: ZN, ARG |
 |
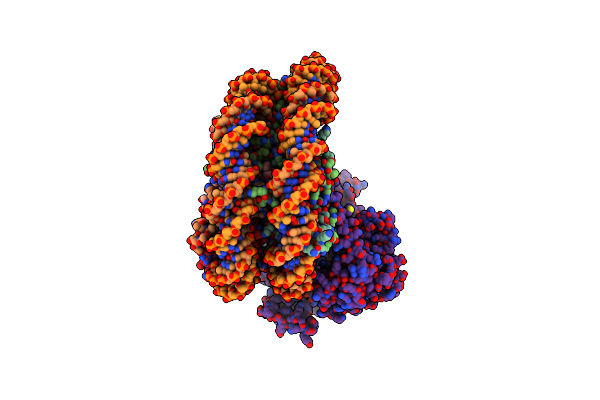
Cryo-Em Structure Of Mmcat1 Bound With Frmlv-Rbd In The Arginine-Bound Inward-Occluded State
Organism: Mus musculus, Murine leukemia virus
Method: ELECTRON MICROSCOPY Release Date: 2025-07-02 Classification: MEMBRANE PROTEIN Ligands: NAG, Y01, ARG |
 |
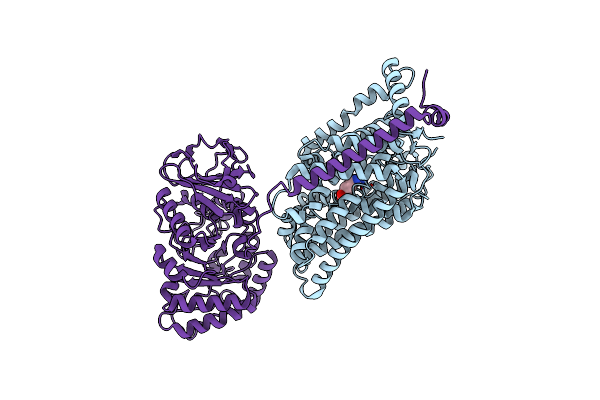
Organism: Xenopus laevis, Synthetic construct, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-05-07 Classification: nuclear protein/DNA Ligands: ARG |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-04-23 Classification: MEMBRANE PROTEIN Ligands: ARG |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-04-02 Classification: MEMBRANE PROTEIN Ligands: ARG |
 |
Crystal Structure Of Histidine Acetyltransferase With L-Arginine And Coenzyme A Disulfide
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-03-19 Classification: TRANSFERASE Ligands: 5NG, GOL, ARG |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-03-12 Classification: MEMBRANE PROTEIN Ligands: ARG |
 |
Organism: Mycolicibacterium smegmatis mc2 155
Method: X-RAY DIFFRACTION Resolution:1.28 Å Release Date: 2024-12-11 Classification: PROTEIN BINDING Ligands: TYR, ARG, MG, GOL, EDO, PGE, NI |
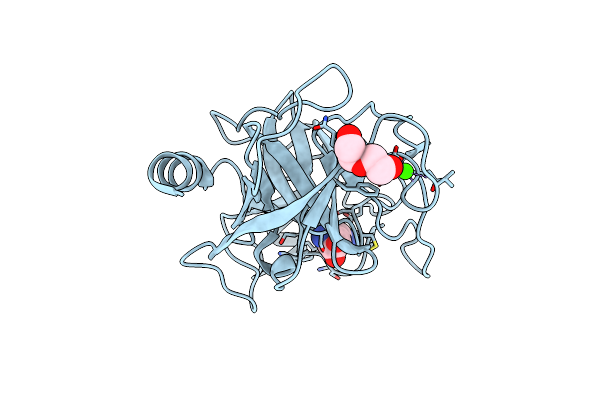 |
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2024-12-04 Classification: HYDROLASE/INHIBITOR Ligands: PEG, CL, CA, ARG |
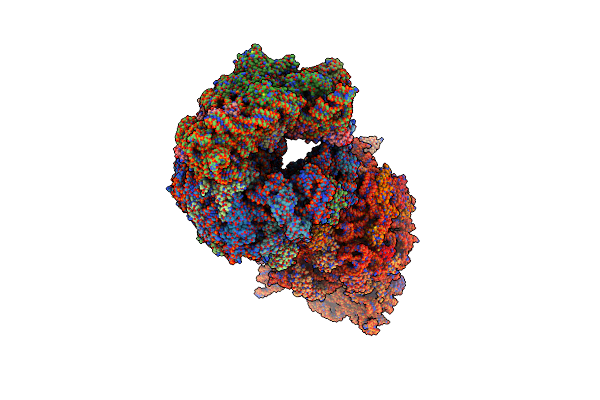 |
Crystal Structure Of The Wild-Type Thermus Thermophilus 70S Ribosome In Complex With Lasso Peptide Lariocidin And Protein Y At 2.60A Resolution
Organism: Escherichia coli k-12, Paenibacillus, Thermus thermophilus hb8
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2024-11-20 Classification: RIBOSOME Ligands: MG, ARG, MPD, ZN, SF4 |
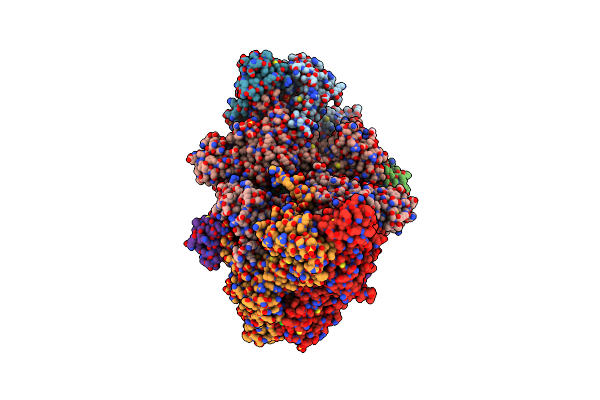 |
Crystal Structure Of Argininosuccinate Lyase From Arabidopsis Thaliana (Atasl) In Complex With Biological Substrate And Products - Argininosuccinate, Argnine And Fumarate
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2024-10-23 Classification: LYASE Ligands: AS1, SO4, CL, FUM, ARG, GOL |
 |
Crystal Structure Of Dimt1 From The Thermophilic Archaeon, Pyrococcus Horikoshii
Organism: Pyrococcus horikoshii ot3
Method: X-RAY DIFFRACTION Resolution:2.01 Å Release Date: 2024-08-28 Classification: TRANSFERASE Ligands: ZN, SO3, GOL, PEG, EDO, ACT, ARG, EPE |
 |
Crystal Structure Of Dimt1 In Complex With 5'-Methylthioadenosine And Adenosine From Pyrococcus Horikoshii
Organism: Pyrococcus horikoshii ot3
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2024-08-28 Classification: TRANSFERASE Ligands: MTA, ADN, ZN, CL, SO3, GOL, EDO, ACT, ARG, PEG |
 |
Crystal Structure Of Dimt1 In Complex With Adenosylornithine (Sfg) From Pyrococcus Horikoshii
Organism: Pyrococcus horikoshii ot3
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2024-08-28 Classification: TRANSFERASE Ligands: SFG, ZN, CL, PEG, ARG, SO3, EDO |
 |
Crystal Structure Of Dimt1 In Complex With S-Adenosyl-L-Homocysteine (Sah) From Pyrococcus Horikoshii
Organism: Pyrococcus horikoshii ot3
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2024-08-28 Classification: TRANSFERASE Ligands: SAH, ZN, SO3, EDO, GOL, PEG, PGE, ARG |

