Search Count: 136
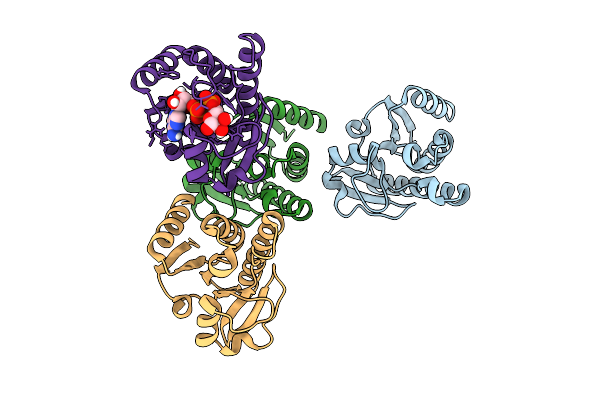 |
Crystal Structure Of Chikungunya Virus Nsp3 Macrodomain D31N Mutant (P41 Crystal Form) In Complex With Adp-Ribose
Organism: Chikungunya virus
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: VIRAL PROTEIN,HYDROLASE Ligands: APR |
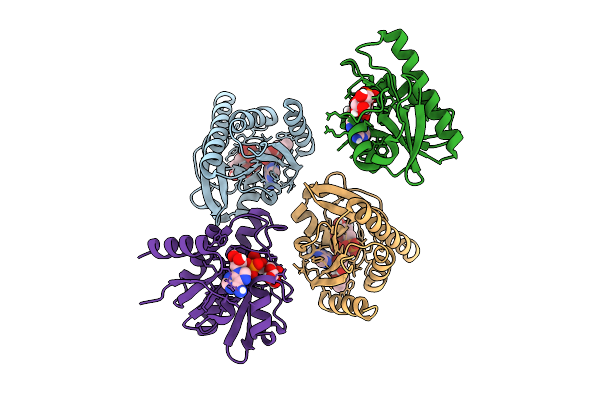 |
Crystal Structure Of Chikungunya Virus Nsp3 Macrodomain N24A Mutant (P31 Crystal Form) In Complex With Adp-Ribose
Organism: Chikungunya virus
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: VIRAL PROTEIN,HYDROLASE Ligands: APR, AR6, IPA |
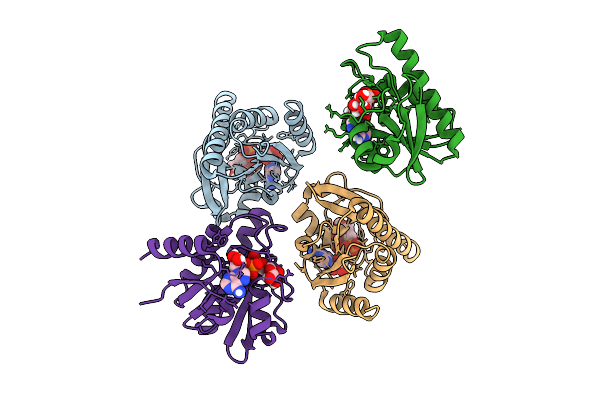 |
Crystal Structure Of Chikungunya Virus Nsp3 Macrodomain N24A D31N Double Mutant (P31 Crystal Form) In Complex With Adp-Ribose
Organism: Chikungunya virus
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: VIRAL PROTEIN,HYDROLASE Ligands: APR, AR6 |
 |
Crystal Structure Of Nad-Dependent Methanol Dehydrogenase 2 From Bacillus Methanolicus Mga3 In Complex With Nad+
Organism: Bacillus methanolicus mga3
Method: X-RAY DIFFRACTION Release Date: 2025-09-03 Classification: OXIDOREDUCTASE Ligands: MN, NAD, APR |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-08-06 Classification: PROTEIN TRANSPORT Ligands: APR |
 |
Crystal Structure Of Methanol Dehydrogenase2 From Bacillus Methanolicus Complexed With An Inhibitor
Organism: Bacillus methanolicus mga3
Method: X-RAY DIFFRACTION Release Date: 2025-06-11 Classification: OXIDOREDUCTASE Ligands: MN, APR |
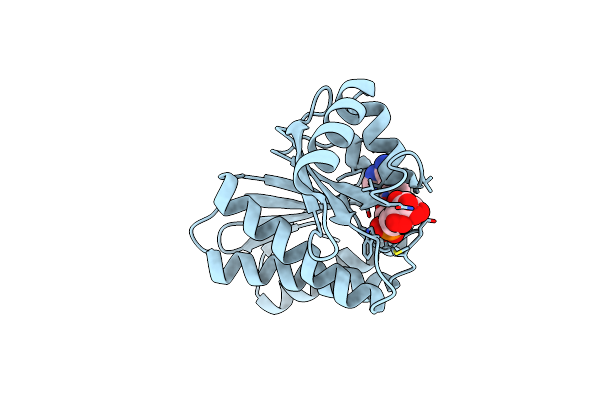 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2025-05-28 Classification: TRANSFERASE Ligands: APR, AR6 |
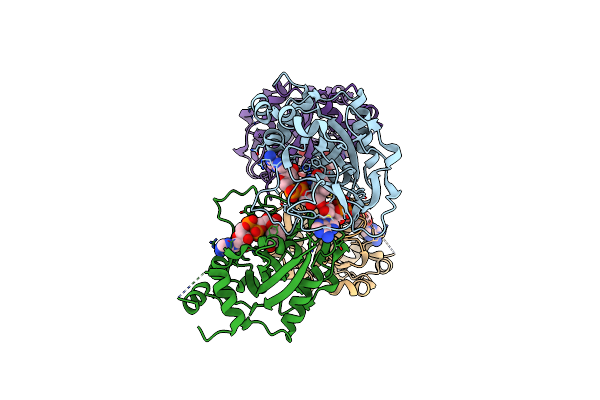 |
Human Parp1 Art Domain Bound To Nad+ Analog Benzamide Adenine Dinucleotide And Adp-Ribose
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2025-02-05 Classification: TRANSFERASE Ligands: DQV, APR |
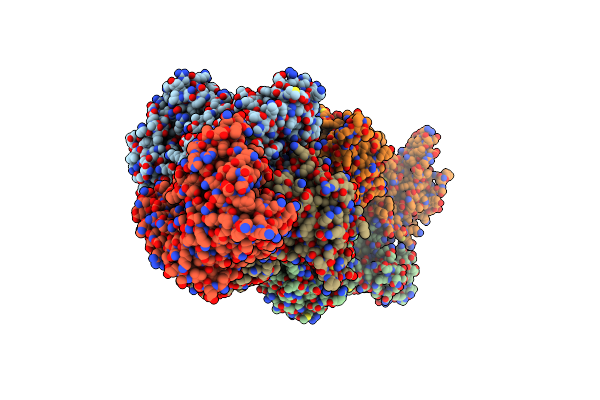 |
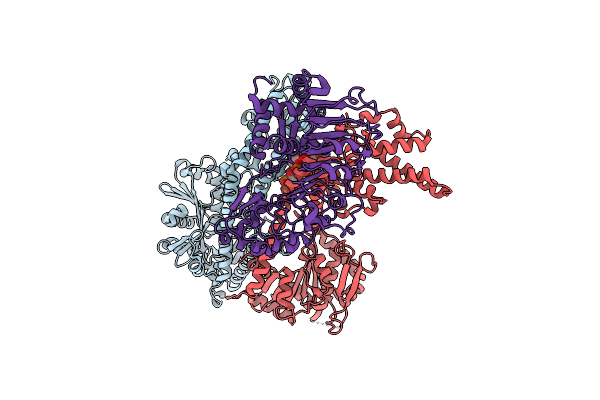
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:2.49 Å Release Date: 2024-12-11 Classification: PLANT PROTEIN/HYDROLASE Ligands: APR, ATP |
 |
Organism: Arabidopsis thaliana
Method: ELECTRON MICROSCOPY Release Date: 2024-12-11 Classification: PLANT PROTEIN/HYDROLASE Ligands: APR, ATP |
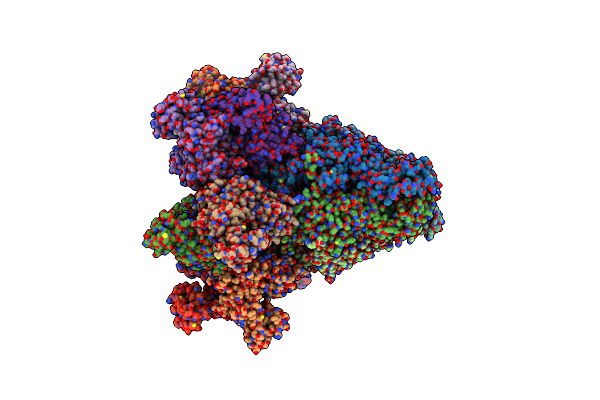 |
Organism: Staphylococcus aureus
Method: ELECTRON MICROSCOPY Release Date: 2024-11-06 Classification: IMMUNE SYSTEM Ligands: APR |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-08-14 Classification: MEMBRANE PROTEIN Ligands: APR |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-08-14 Classification: MEMBRANE PROTEIN Ligands: APR, AR6 |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-08-14 Classification: MEMBRANE PROTEIN Ligands: APR, DAT, ZN, CA |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-08-14 Classification: MEMBRANE PROTEIN Ligands: APR, ZN |
 |
Cryo-Em Structure Of Trpm2 Chanzyme In The Presence Of Magnesium And Adp-Ribose, Open State
Organism: Salpingoeca rosetta
Method: ELECTRON MICROSCOPY Release Date: 2024-05-15 Classification: MEMBRANE PROTEIN Ligands: APR, MG, CLR |
 |
Cryo-Em Structure Of Trpm2 Chanzyme In The Presence Of Magnesium And Adp-Ribose, Closed State
Organism: Salpingoeca rosetta
Method: ELECTRON MICROSCOPY Release Date: 2024-05-15 Classification: MEMBRANE PROTEIN Ligands: APR, MG, CLR |
 |
Cryo-Em Structure Of Trpm2 Chanzyme In The Presence Of Magnesium, Adp-Ribose, Adenosine Monophosphate, And Ribose-5-Phosphate, Closed State
Organism: Salpingoeca rosetta
Method: ELECTRON MICROSCOPY Release Date: 2024-05-15 Classification: MEMBRANE PROTEIN Ligands: RP5, AMP, APR, MG, CLR |
 |
Cryo-Em Structure Of Trpm2 Chanzyme (E1114A) In The Presence Of Magnesium And Adp-Ribose, Open State
Organism: Salpingoeca rosetta
Method: ELECTRON MICROSCOPY Release Date: 2024-05-15 Classification: MEMBRANE PROTEIN Ligands: APR, MG, CLR |
 |
Cryo-Em Structure Of Trpm2 Chanzyme (E1114A) In The Presence Of Magnesium And Adp-Ribose, Closed State
Organism: Salpingoeca rosetta
Method: ELECTRON MICROSCOPY Release Date: 2024-05-15 Classification: MEMBRANE PROTEIN Ligands: APR, MG, CLR |

