Search Count: 31
 |
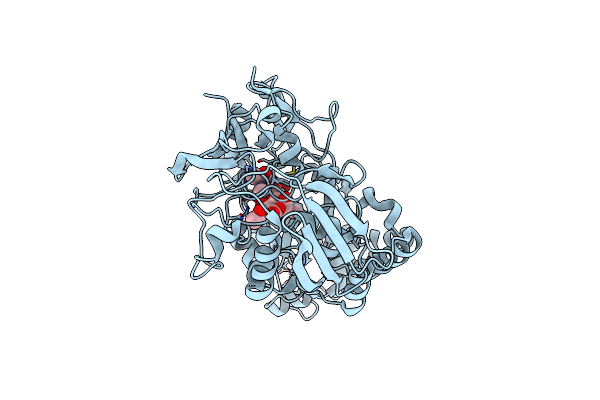
Crystal Structure Of Chitin Oligosaccharide Binding Protein From Vibrio Cholera.
Organism: Vibrio cholerae
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2024-01-03 Classification: SUGAR BINDING PROTEIN Ligands: MG |
 |
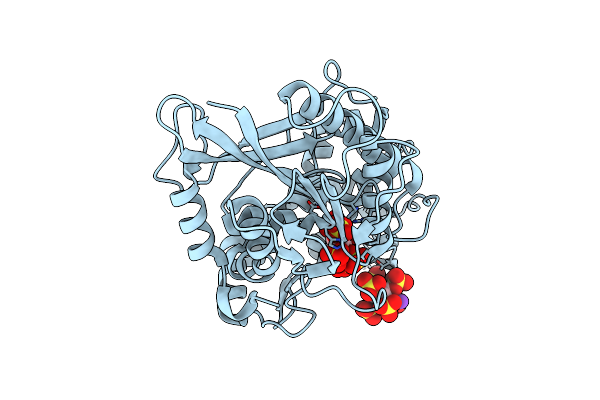
Crystal Structure Of Chitin Oligosaccharide Binding Protein From Vibrio Cholera In Complex With Chitotriose.
Organism: Vibrio cholerae
Method: X-RAY DIFFRACTION Resolution:1.22 Å Release Date: 2024-01-03 Classification: SUGAR BINDING PROTEIN Ligands: MG |
 |
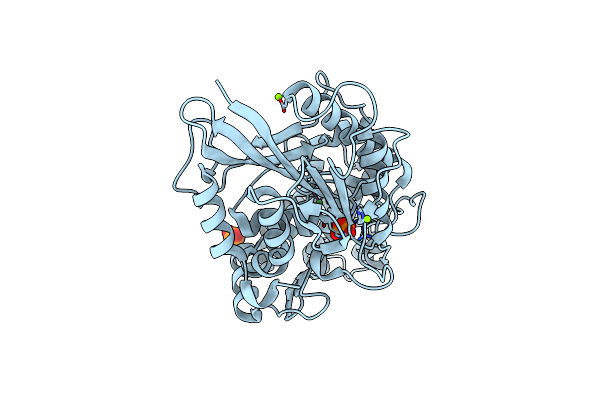
Escherichia Coli Periplasmic Phytase Appa D304E Mutant, Complex With Myo-Inositol Hexakissulfate
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2022-06-29 Classification: HYDROLASE Ligands: IHS, K |
 |
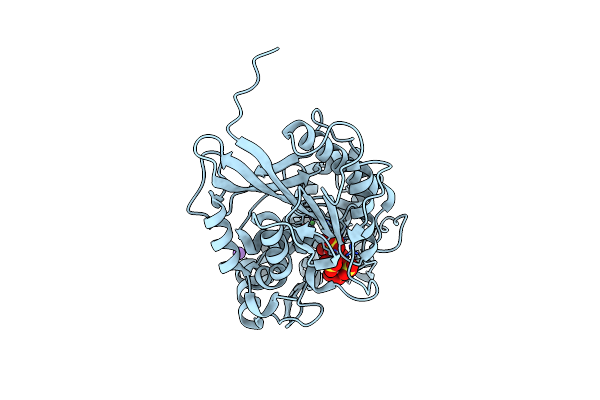
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2022-03-16 Classification: HYDROLASE Ligands: PO4, NI, MG |
 |
Escherichia Coli Periplasmic Phytase Appa, Complex With Myo-Inositol Hexakissulfate
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.72 Å Release Date: 2022-03-16 Classification: HYDROLASE Ligands: IHS, NI, K |
 |
Escherichia Coli Periplasmic Phytase Appa D304A Mutant, Complex With Myo-Inositol Hexakissulfate
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.41 Å Release Date: 2022-03-16 Classification: HYDROLASE Ligands: IHS, GOL, PO4, MES |
 |
Escherichia Coli Periplasmic Phytase Appa D304A,T305E Mutant, Complex With Myo-Inositol Hexakissulfate
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.42 Å Release Date: 2022-03-16 Classification: HYDROLASE Ligands: IHS, GOL, PO4, MES |
 |
Escherichia Coli Periplasmic Phytase Appa T305E Mutant, Complex With Myo-Inositol Hexakissulfate
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.86 Å Release Date: 2022-03-16 Classification: HYDROLASE Ligands: IHS, NI |
 |
Escherichia Coli Periplasmic Phytase Appa D304A Mutant, Phosphohistidine Intermediate
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2022-03-16 Classification: HYDROLASE Ligands: NI |
 |
Phenylalanine Ammonia-Lyase (Pal) From Petroselinum Crispum Complexed With S-Appa
Organism: Petroselinum crispum
Method: X-RAY DIFFRACTION Release Date: 2019-06-26 Classification: LYASE Ligands: CV2 |
 |
Crystal Structure Of A Putative Peptide Binding Protein Appa From Clostridium Difficile
Organism: Clostridioides difficile
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2019-04-10 Classification: PEPTIDE BINDING PROTEIN Ligands: IOD, SO4 |
 |
Organism: Bacillus subtilis subsp. spizizenii
Method: X-RAY DIFFRACTION Resolution:1.97 Å Release Date: 2019-03-13 Classification: LYASE Ligands: LJS |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.07 Å Release Date: 2015-05-06 Classification: HYDROLASE/HYDROLASE Inhibitor Ligands: NI, IHS |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.27 Å Release Date: 2014-07-23 Classification: SIGNALING PROTEIN |
 |
Organism: Rhodobacter sphaeroides
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2014-02-12 Classification: TRANSCRIPTION Ligands: SO4, MES |
 |
Organism: Rhodobacter sphaeroides
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2013-09-18 Classification: METAL BINDING PROTEIN |
 |
Dark-State Structure Of Appa C20S Without The Cys-Rich Region From Rb. Sphaeroides
Organism: Rhodobacter sphaeroides
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2013-06-05 Classification: FLAVOPROTEIN,SIGNALING PROTEIN Ligands: FMN, CL |
 |
Dark-State Structure Of Appa Wild-Type Without The Cys-Rich Region From Rb. Sphaeroides
Organism: Rhodobacter sphaeroides
Method: X-RAY DIFFRACTION Resolution:3.50 Å Release Date: 2013-06-05 Classification: FLAVOPROTEIN,SIGNALING PROTEIN Ligands: FMN |
 |
Organism: Rhodobacter sphaeroides
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2013-06-05 Classification: TRANSCRIPTION |
 |
Organism: Rhodobacter sphaeroides
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2013-06-05 Classification: FLAVOPROTEIN/TRANSCRIPTION |

