Search Count: 21
All
Selected
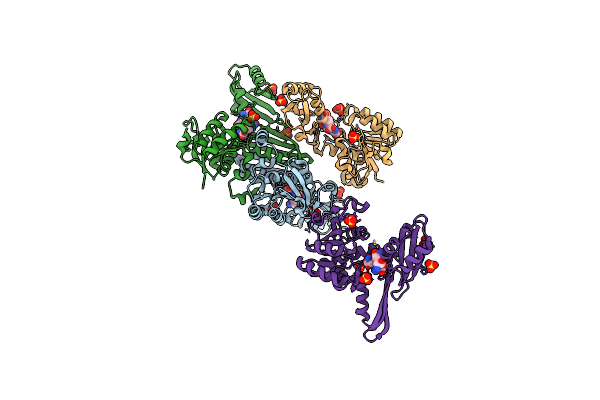 |
Organism: Proteus vulgaris
Method: X-RAY DIFFRACTION Resolution:3.02 Å Release Date: 2024-02-07 Classification: OXIDOREDUCTASE Ligands: API, SO4 |
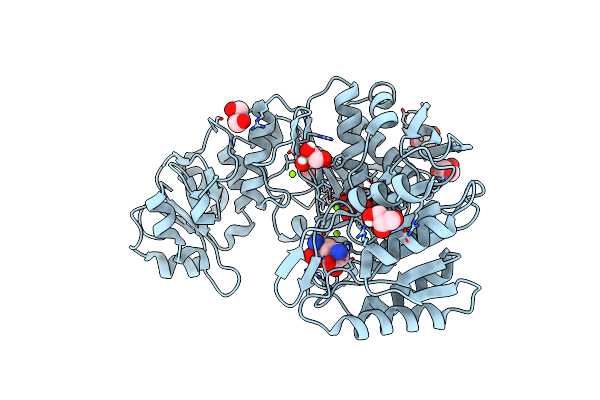 |
Crystal Structure Of Mycobacterium Thermoresistibile Mure In Complex With Adp And 2,6-Diaminopimelic Acid
Organism: Mycolicibacterium thermoresistibile
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2023-04-05 Classification: LIGASE Ligands: MG, GOL, ADP, API |
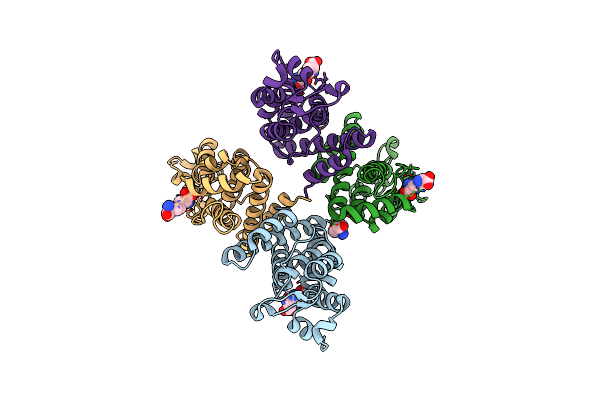 |
Organism: Salmonella typhi
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2020-01-29 Classification: PROTEIN TRANSPORT Ligands: API, TRS |
 |
Crystal Structure Of Dape In Complex With The Products (Succinic Acid And Diaminopimelic Acid)
Organism: Haemophilus influenzae
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2018-07-25 Classification: HYDROLASE Ligands: SIN, API, ZN |
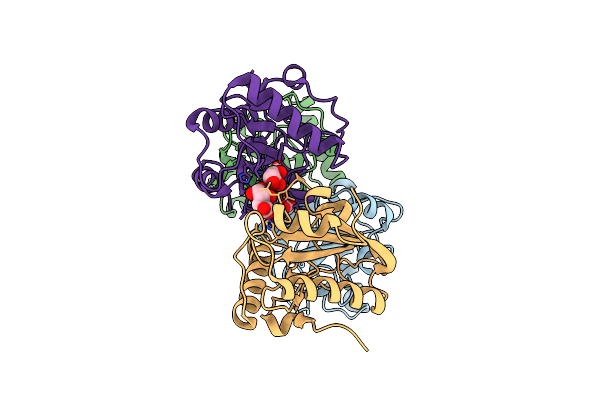 |
Crystal Structure Of The Ternary Complex Of Peptidoglycan Recognition Protein (Pgrp-S) With Tartaric Acid, Ribose And 2,6-Diaminopimelic Acid At 2.11 A Resolution
Organism: Camelus dromedarius
Method: X-RAY DIFFRACTION Resolution:2.11 Å Release Date: 2018-07-25 Classification: IMMUNE SYSTEM Ligands: EDO, API, GOL, TLA, RIB |
 |
Crystal Structure Of The Peptidoglycan-Associated Lipoprotein (Pal) From Burkholderia Cepacia In Complex With Dap
Organism: Burkholderia cenocepacia (strain atcc baa-245 / dsm 16553 / lmg 16656 / nctc 13227 / j2315 / cf5610)
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2017-08-16 Classification: MEMBRANE PROTEIN Ligands: API |
 |
Crystal Structure Of Dapf From Corynebacterium Glutamicum In Complex With D,L-Diaminopimelate
Organism: Corynebacterium glutamicum (strain atcc 13032 / dsm 20300 / jcm 1318 / lmg 3730 / ncimb 10025)
Method: X-RAY DIFFRACTION Resolution:2.59 Å Release Date: 2016-11-02 Classification: ISOMERASE Ligands: API |
 |
Crystal Structure Of Apo Cell Shape Determinant Protein Csd4 From Helicobacter Pylori
Organism: Helicobacter pylori
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2014-12-24 Classification: HYDROLASE Ligands: IOD, API |
 |
Crystal Structure Of Product Bound Cell Shape Determinant Protein Csd4 From Helicobacter Pylori
Organism: Helicobacter pylori
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2014-12-24 Classification: HYDROLASE Ligands: ZN, IOD, API, NA |
 |
Crystal Structure Of Cell Shape Determinant Protein Csd4 Gln46His Variant From Helicobacter Pylori
Organism: Helicobacter pylori
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2014-12-24 Classification: HYDROLASE Ligands: IOD, ZN, PO4, API |
 |
Structural Analysis Of The Mdap-Bound Form Of Helicobacter Pylori Csd4, A D,L-Carboxypeptidase
Organism: Helicobacter pylori
Method: X-RAY DIFFRACTION Resolution:1.41 Å Release Date: 2014-11-05 Classification: HYDROLASE Ligands: CA, API, GOL |
 |
Structural Analysis Of The Zn-Form I Of Helicobacter Pylori Csd4, A D,L-Carboxypeptidase
Organism: Helicobacter pylori
Method: X-RAY DIFFRACTION Resolution:2.62 Å Release Date: 2014-11-05 Classification: HYDROLASE Ligands: ZN, CA, API |
 |
Structural Analysis Of The Zn-Form Ii Of Helicobacter Pylori Csd4, A D,L-Carboxypeptidase
Organism: Helicobacter pylori
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2014-11-05 Classification: HYDROLASE Ligands: ZN, CA, API |
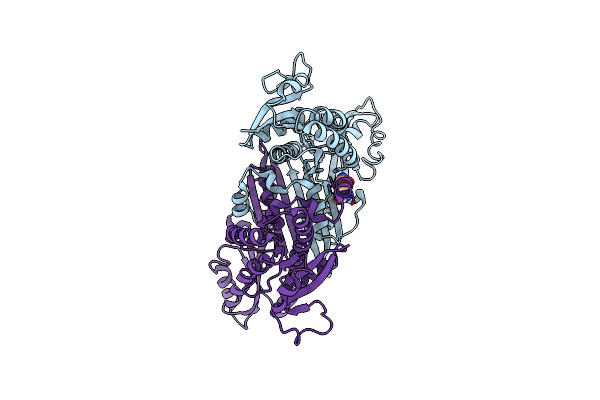 |
Crystal Structure Of Meso-Dapdh Q154L/T173I/R199M/P248S/H249N/N276S Mutant With Dap Of From Clostridium Tetani E88
Organism: Clostridium tetani
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2014-08-20 Classification: OXIDOREDUCTASE Ligands: API |
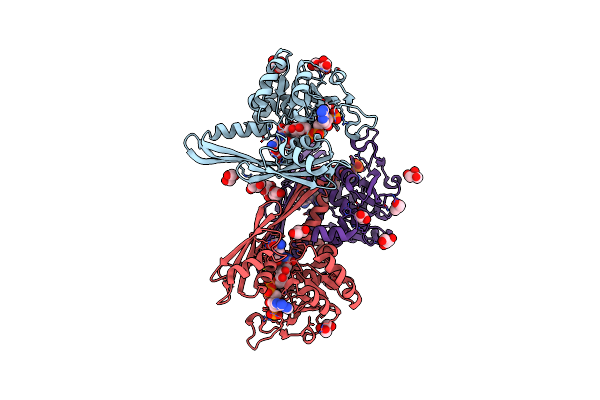 |
Crystal Structure Of Meso-Diaminopimelate Dehydrogenase From Symbiobacterium Thermophilum Co-Crystallized With Nadp+ And Dap
Organism: Symbiobacterium thermophilum
Method: X-RAY DIFFRACTION Resolution:2.12 Å Release Date: 2014-03-26 Classification: OXIDOREDUCTASE Ligands: API, GOL, NAP |
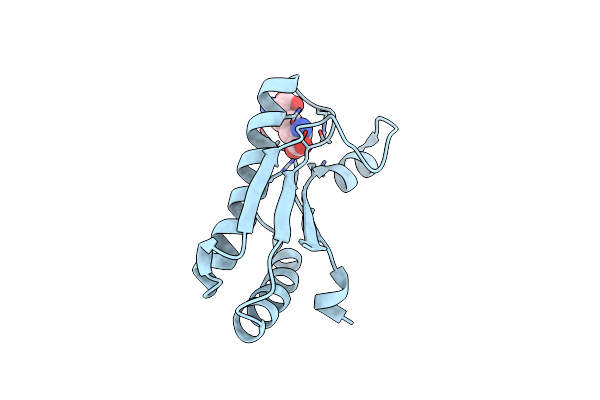 |
Crystal Structure Of Peptidoglycan-Associated Lipoprotein From Acinetobacter Baumannii
Organism: Acinetobacter baumannii
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2013-07-24 Classification: PEPTIDE BINDING PROTEIN Ligands: API |
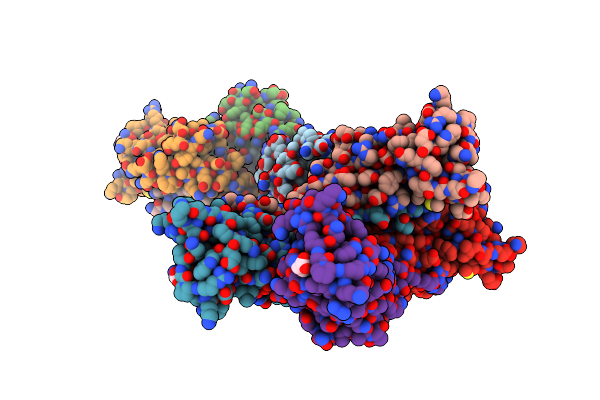 |
Crystal Structure Of Ompa Peptidoglycan-Binding Domain From Acinetobacter Baumannii
Organism: Acinetobacter baumannii
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2013-07-24 Classification: PEPTIDE BINDING PROTEIN Ligands: API, SRT |
 |
Crystal Structure Of C-Lobe Of Bovine Lactoferrin Complexed With Diaminopimelic Acid At 1.46 A Resolution
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:1.46 Å Release Date: 2012-01-18 Classification: METAL BINDING PROTEIN Ligands: NAG, FE, ZN, CO3, SO4, API |
 |
Crystal Structure Of Ompa-Like Domain From Acinetobacter Baumannii In Complex With Diaminopimelate
Organism: Acinetobacter baumannii
Method: X-RAY DIFFRACTION Resolution:1.79 Å Release Date: 2011-10-26 Classification: MEMBRANE PROTEIN,PEPTIDE BINDING PROTEIN Ligands: API |
 |
Structure Of Mure The Udp-N-Acetylmuramyl Tripeptide Synthetase From E. Coli
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2001-09-13 Classification: LIGASE Ligands: CL, UAG, API |

