Search Count: 145
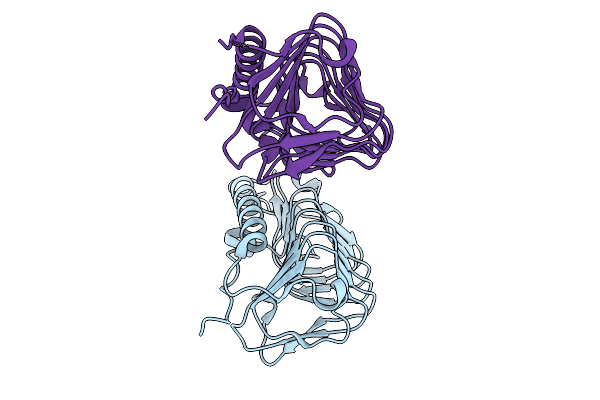 |
Crystal Structure Of Ice Binding Protein From Candidatus Cryosericum Odellii Smc5
Organism: Candidatus cryosericum odellii
Method: X-RAY DIFFRACTION Release Date: 2025-09-03 Classification: RECOMBINATION |
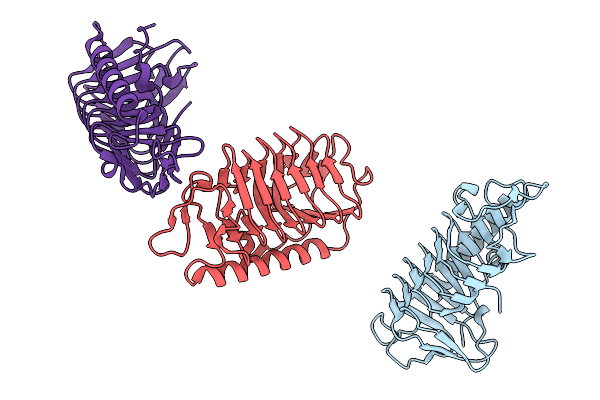 |
Crystal Structure Of Ice Binding Protein From Candidatus Cryosericum Odellii Smc5
Organism: Candidatus cryosericum odellii
Method: X-RAY DIFFRACTION Release Date: 2025-09-03 Classification: RECOMBINATION |
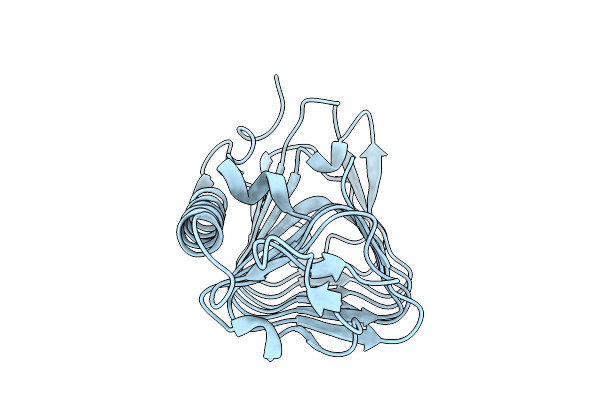 |
Crystal Structure Of An Ice-Binding Protein From Candidatus Cryosericum Odellii
Organism: Candidatus cryosericum odellii
Method: X-RAY DIFFRACTION Release Date: 2025-09-03 Classification: PROTEIN BINDING |
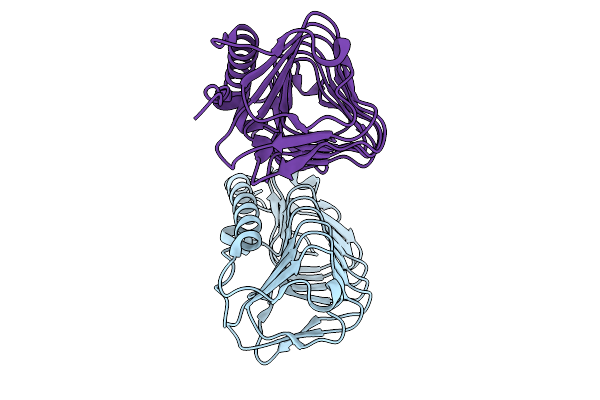 |
Crystal Structure Of Engineered Ice-Binding Protein (M74I And A97V) From Candidatus Cryosericum Odellii Strain Smc5_169
Organism: Candidatus cryosericum odellii
Method: X-RAY DIFFRACTION Release Date: 2025-09-03 Classification: PROTEIN BINDING |
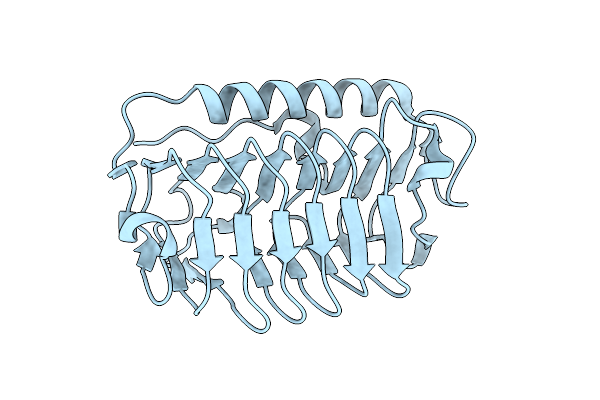 |
Organism: Flavobacterium frigoris ps1
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2024-11-13 Classification: ANTIFREEZE PROTEIN |
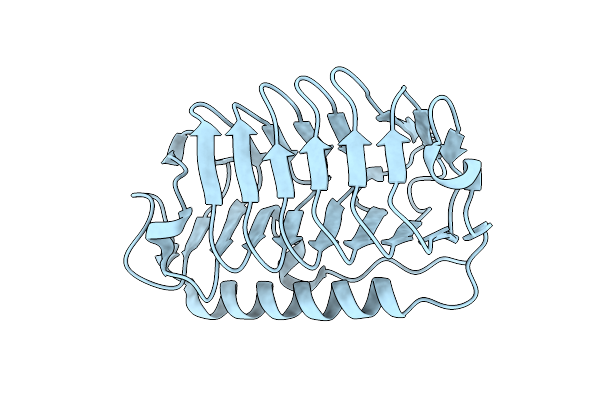 |
Organism: Flavobacterium frigoris ps1
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2024-11-13 Classification: ANTIFREEZE PROTEIN |
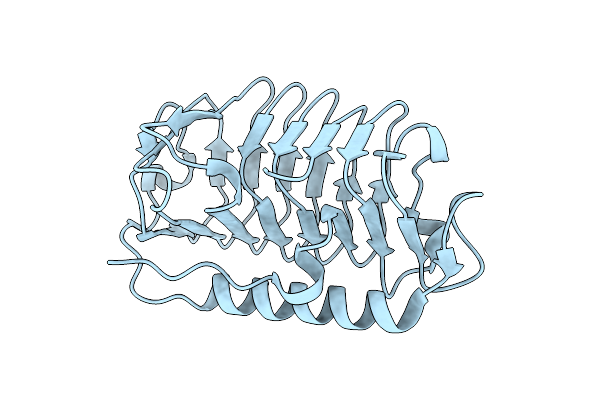 |
Organism: Flavobacterium frigoris ps1
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2024-11-13 Classification: ANTIFREEZE PROTEIN |
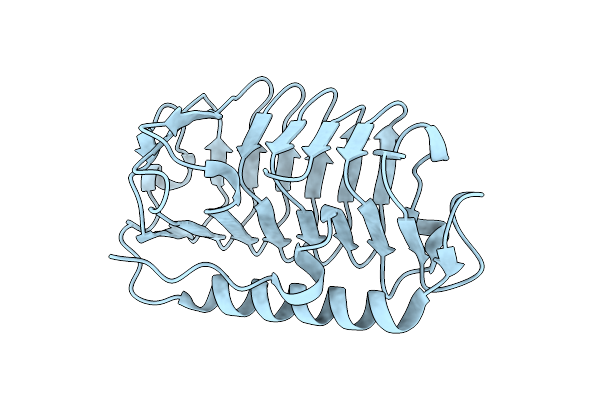 |
Organism: Flavobacterium frigoris ps1
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2024-11-13 Classification: ANTIFREEZE PROTEIN |
 |
Structure Of A Type Iii Antifreeze Protein Isoform Hplc12 Re-Refined Using Standard Protocols
Organism: Zoarces americanus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2024-07-17 Classification: ANTIFREEZE PROTEIN |
 |
Organism: Marine sediment metagenome
Method: X-RAY DIFFRACTION Resolution:3.05 Å Release Date: 2023-09-13 Classification: ANTIFREEZE PROTEIN Ligands: PG4 |
 |
Crystal Structure Of The Pseudaminic Acid Synthase Psei From Campylobacter Jejuni
Organism: Campylobacter jejuni
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2022-11-09 Classification: BIOSYNTHETIC PROTEIN Ligands: MN |
 |
Organism: Zoarces americanus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2022-04-20 Classification: ANTIFREEZE PROTEIN |
 |
Organism: Flavobacterium frigoris ps1
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2022-03-02 Classification: ANTIFREEZE PROTEIN Ligands: CL |
 |
Organism: Typhula ishikariensis
Method: X-RAY DIFFRACTION Resolution:1.72 Å Release Date: 2021-11-03 Classification: ANTIFREEZE PROTEIN Ligands: MG |
 |
Organism: Typhula ishikariensis
Method: X-RAY DIFFRACTION Resolution:1.54 Å Release Date: 2021-10-27 Classification: ANTIFREEZE PROTEIN Ligands: SO4 |
 |
Organism: Marinomonas primoryensis, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2021-09-01 Classification: PROTEIN BINDING Ligands: 1PE, CA |
 |
Organism: Marinomonas primoryensis, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2021-09-01 Classification: PROTEIN BINDING Ligands: CA, NA, 1PE |
 |
Organism: Marinomonas primoryensis, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2021-09-01 Classification: PROTEIN BINDING Ligands: CA, 1PE |
 |
Organism: Marinomonas primoryensis, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.17 Å Release Date: 2021-09-01 Classification: PROTEIN BINDING Ligands: CA, 1PE |
 |
Fucose-Bound Structure Of Marinomonas Primoryensis Pa14 Carbohydrate-Binding Domain
Organism: Marinomonas primoryensis
Method: X-RAY DIFFRACTION Resolution:0.97 Å Release Date: 2021-06-02 Classification: SUGAR BINDING PROTEIN Ligands: FUL, CA, EDO, FUC |

