Search Count: 3,780
 |
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: LIGASE/ANTIBIOTIC Ligands: XMP, A1L67 |
 |
Organism: Enterococcus casseliflavus
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: ANTIBIOTIC Ligands: A1I7W, DMS |
 |
M. Tuberculosis Clpc1-Ntd Complexed With A Click Chemistry Analog Of Rufomycin
Organism: Mycobacterium tuberculosis, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: ANTIBIOTIC |
 |
Organism: Vibrio cholerae o1 biovar el tor str. n16961
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: HYDROLASE Ligands: NI, BB2 |
 |
Organism: Vibrio cholerae o1 biovar el tor str. n16961
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: HYDROLASE Ligands: BB2, NI |
 |
Organism: Thermothelomyces thermophilus, Photorhabdus khanii
Method: ELECTRON MICROSCOPY Release Date: 2025-12-03 Classification: MEMBRANE PROTEIN/ANTIBIOTIC Ligands: ERG |
 |
An Antibiotic Biosynthesis Monooxygenase Family Protein From Streptomyces Sp. Ma37
Organism: Streptomyces sp. ma37
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: ANTIBIOTIC |
 |
Crystal Structure Of A Polyketide Abm/Scha-Like Domain-Containing Protein Whie-Orfi From Streptomyces Coelicolor
Organism: Streptomyces coelicolor
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: ANTIBIOTIC |
 |
Organism: Staphylococcus aureus
Method: ELECTRON MICROSCOPY Release Date: 2025-11-26 Classification: MEMBRANE PROTEIN |
 |
Organism: Staphylococcus aureus
Method: ELECTRON MICROSCOPY Release Date: 2025-11-26 Classification: MEMBRANE PROTEIN |
 |
Organism: Staphylococcus aureus
Method: ELECTRON MICROSCOPY Release Date: 2025-11-26 Classification: MEMBRANE PROTEIN Ligands: ADP |
 |
Organism: Escherichia coli bw25113
Method: ELECTRON MICROSCOPY Release Date: 2025-11-26 Classification: ANTIBIOTIC |
 |
Organism: Enterococcus casseliflavus
Method: X-RAY DIFFRACTION Release Date: 2025-11-19 Classification: ANTIBIOTIC Ligands: DMS, A1I8X |
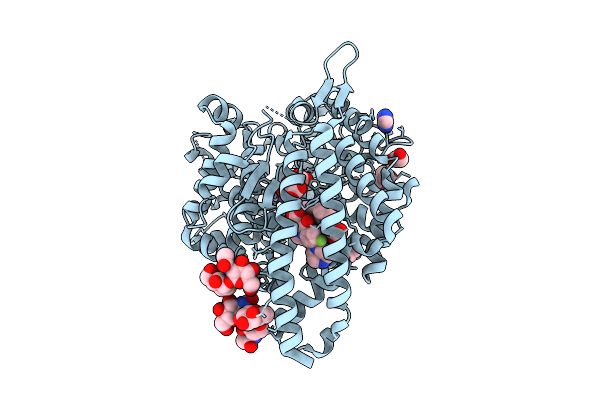 |
Human Angiotensin-1 Converting Enzyme C-Domain In Complex With Ciprofloxacin
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-11-12 Classification: HYDROLASE Ligands: ZN, CL, IMD, NXA, MLI, CPF, EDO, PEG |
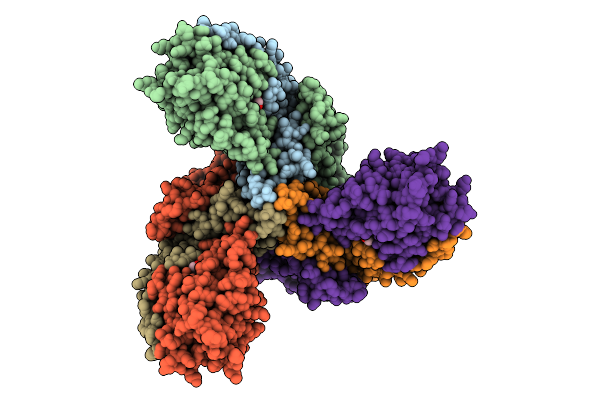 |
Organism: Lactococcus garvieae
Method: ELECTRON MICROSCOPY Release Date: 2025-11-12 Classification: MEMBRANE PROTEIN Ligands: MAN |
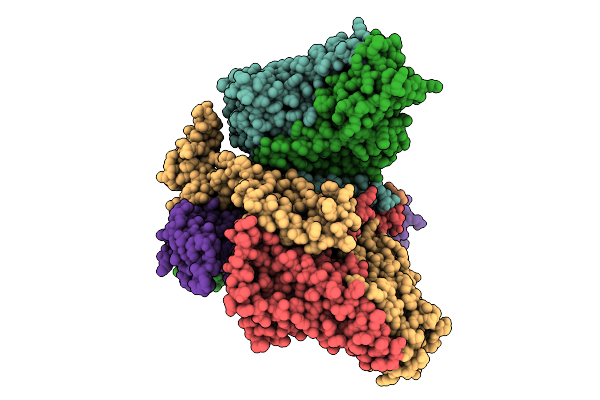 |
Cryo-Em Structure Of The L. Garvieae Man-Pts In Complex With The Bacteriocin Garq
Organism: Lactococcus garvieae
Method: ELECTRON MICROSCOPY Release Date: 2025-11-12 Classification: MEMBRANE PROTEIN Ligands: MAN |
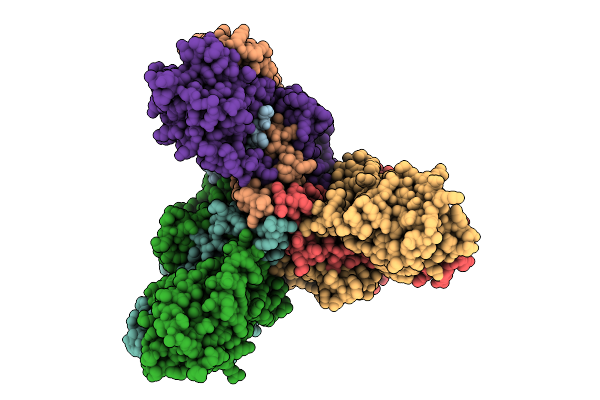 |
Organism: Listeria monocytogenes, Lactococcus garvieae
Method: ELECTRON MICROSCOPY Release Date: 2025-11-12 Classification: MEMBRANE PROTEIN Ligands: MAN |
 |
Organism: Escherichia coli, Streptomyces clavuligerus
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: HYDROLASE/PROTEIN BINDING |
 |
Organism: Escherichia coli (strain k12)
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: PROTEIN TRANSPORT |
 |
Organism: Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: PROTEIN TRANSPORT |

