Search Count: 43
 |
Organism: Mus musculus, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: SIGNALING PROTEIN Ligands: CA |
 |
Organism: Mus musculus, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: SIGNALING PROTEIN Ligands: CA |
 |
Organism: Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2025-02-05 Classification: CELL ADHESION Ligands: CA |
 |
Crystal Structure Of The Hormr-Gain Domains Of Adhesion Gpcr Adgrb2 (Bai2) In The Uncleaved State
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.22 Å Release Date: 2023-05-31 Classification: SIGNALING PROTEIN Ligands: NAG, CAC |
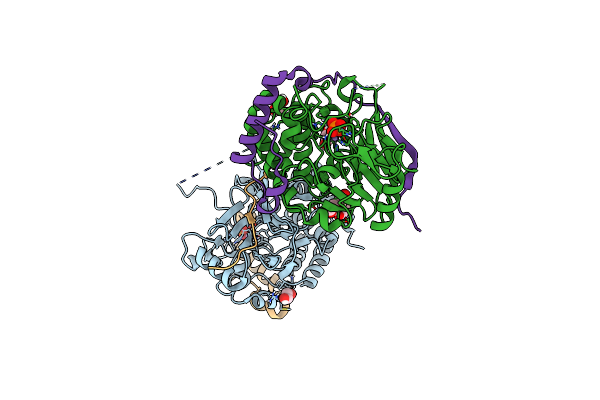 |
Structure Of Pp1-Irsp53 Chimera [Pp1(7-304) + Linker (G/S)X9 + Irsp53(449-465)] Bound To Phactr1 (516-580)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.09 Å Release Date: 2020-09-30 Classification: HYDROLASE Ligands: MN, PO4, EDO, 16P |
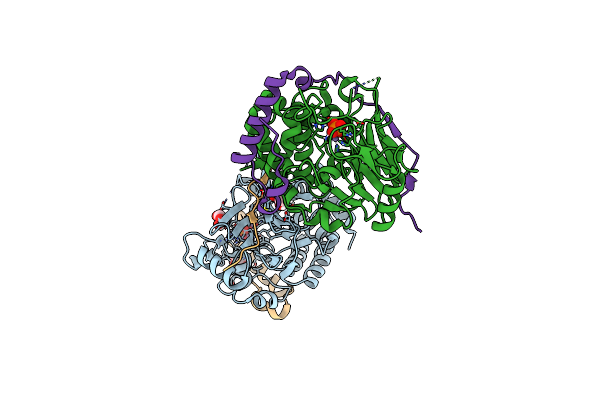 |
Structure Of Pp1-Irsp53 S455E Chimera [Pp1(7-304) + Linker (G/S)X9 + Irsp53(449-465)] Bound To Phactr1 (516-580)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.39 Å Release Date: 2020-09-30 Classification: HYDROLASE Ligands: MN, GOL, PO4 |
 |
Organism: Homo sapiens, Mus musculus
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2019-01-23 Classification: CELL ADHESION |
 |
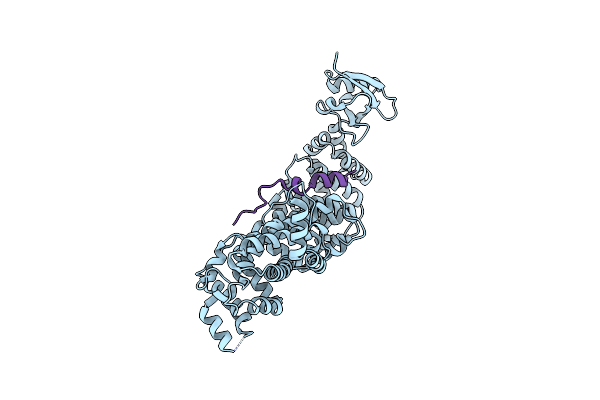
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.39 Å Release Date: 2015-04-22 Classification: PROTEIN BINDING Ligands: MG |
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.03 Å Release Date: 2015-04-22 Classification: PROTEIN BINDING Ligands: MG |
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.38 Å Release Date: 2015-04-15 Classification: PROTEIN BINDING |
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2015-04-15 Classification: PROTEIN BINDING Ligands: CD |
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2015-04-15 Classification: PROTEIN BINDING Ligands: CD |
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:1.46 Å Release Date: 2015-04-15 Classification: PROTEIN BINDING Ligands: CD |
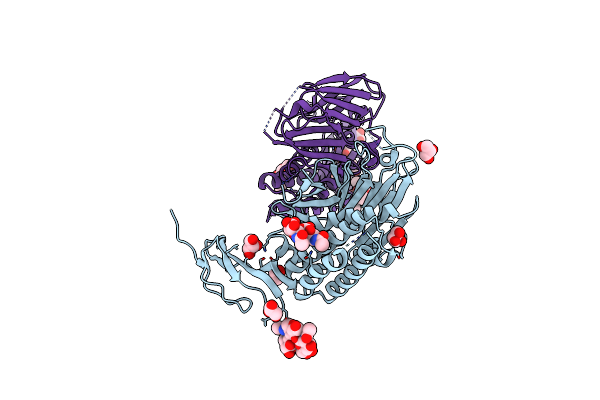 |
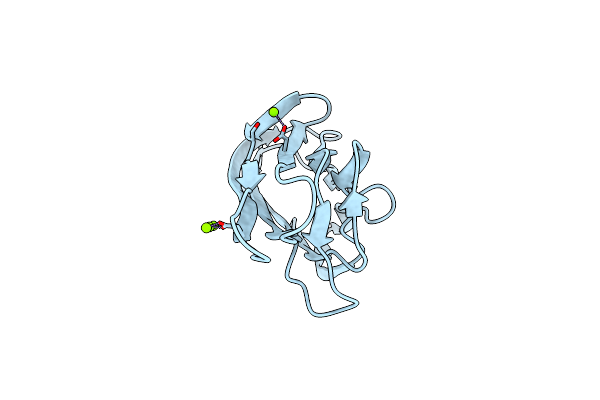
Crystal Structure Of The Gain And Hormr Domains Of Brain Angiogenesis Inhibitor 3 (Bai3)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2012-02-22 Classification: SIGNALING PROTEIN Ligands: GOL, NAG |
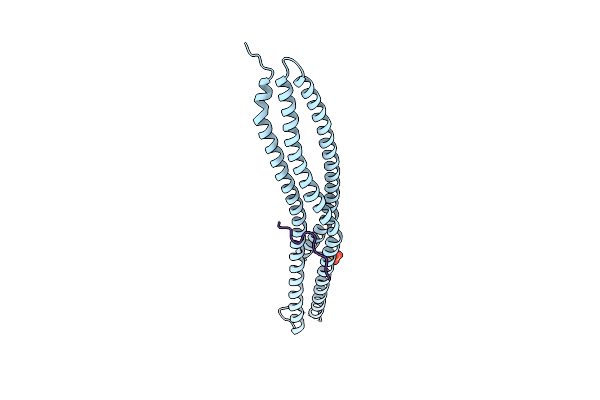 |
Crystal Structure Of The I-Bar Domain Of Irsp53 (Baiap2) In Complex With An Ehec Derived Tir Peptide
Organism: Homo sapiens, Escherichia coli o157\:h7
Method: X-RAY DIFFRACTION Resolution:2.11 Å Release Date: 2011-09-07 Classification: SIGNALING PROTEIN Ligands: SO4 |
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2011-07-13 Classification: PROTEIN BINDING Ligands: GOL |
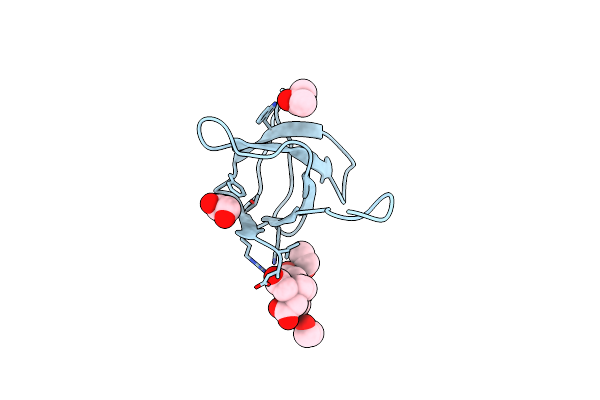 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2011-05-04 Classification: PROTEIN BINDING Ligands: EDT, EDO, IPA |
 |
1H, 13C, And 15N Chemical Shift Assignments For Irtks-Sh3 And Espfu-R47 Complex
Organism: Homo sapiens, Escherichia coli
Method: SOLUTION NMR Release Date: 2010-11-17 Classification: PROTEIN BINDING |
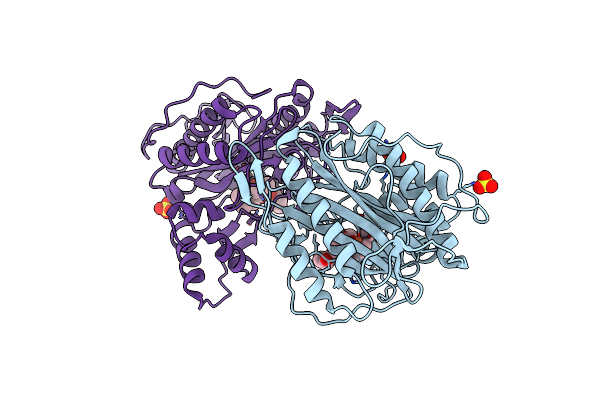 |
Crystal Structure Of An Encephalitozoon Cuniculi Methionine Aminopeptidase Type 2 With Angiogenesis Inhibitor Fumagillin Bound
Organism: Encephalitozoon cuniculi
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2009-01-13 Classification: HYDROLASE Ligands: FE, FUG, SO4 |
 |
Crystal Structure Of An Encephalitozoon Cuniculi Methionine Aminopeptidase Type 2 With Angiogenesis Inhibitor Tnp470 Bound
Organism: Encephalitozoon cuniculi
Method: X-RAY DIFFRACTION Resolution:2.89 Å Release Date: 2009-01-13 Classification: HYDROLASE Ligands: FE, TN4, SO4 |

